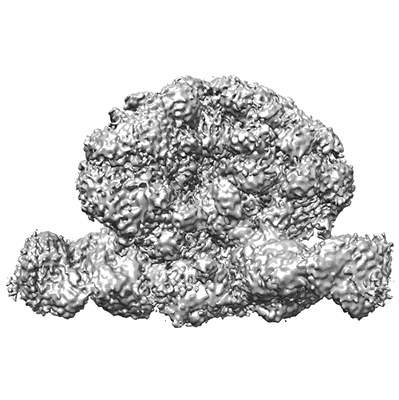
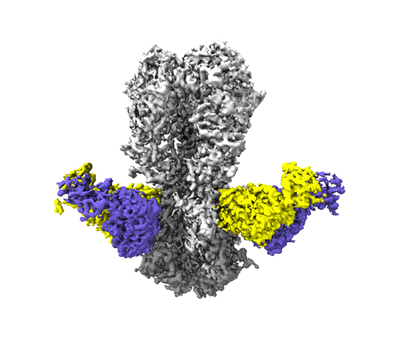
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 8fr6
Q-score: 0.528
Sastry M, Changela A, Gorman J, Xu K, Chuang GY, Shen CH, Cheng C, Geng H, O'Dell S, Ou L, Rawi R, Reveiz M, Stewart-Jones GBE, Wang S, Zhang B, Zhou T, Biju A, Chambers M, Chen X, Corrigan AR, Lin BC, Louder MK, McKee K, Nazzari AF, Olia AS, Parchment DK, Sarfo EK, Stephens T, Stuckey J, Tsybovsky Y, Verardi R, Wang Y, Zheng CY, Chen Y, Doria-Rose NA, McDermott AB, Mascola JR, Kwong PD
J Virol (2023) 97 pp. e0160422-e0160422 [ Pubmed: 37098956 DOI: doi:10.1128/jvi.01604-22 ]
- Antibody fab (Complex from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Envelope glycoprotein gp120 (52 kDa, Protein from Human immunodeficiency virus 1)
- Complex of antibody fab vfp53.02 with hiv-1 env trimer bg505 ds-sosip (Complex)
- Vfp53.02 fab heavy chain (24 kDa, Protein from Mus musculus)
- Hiv-1 env trimer bg505 ds-sosip (Complex from Human immunodeficiency virus 1)
- Vfp53.02 fab light chain (24 kDa, Protein from Mus musculus)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
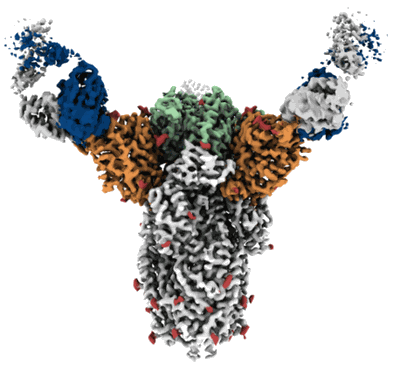
Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Resolution: 2.83 Å
EM Method: Single-particle
Fitted PDBs: 7tb8
Q-score: 0.41
Zhou T, Wang L, Misasi J, Pegu A, Zhang Y, Harris DR, Olia AS, Talana CA, Yang ES, Chen M, Choe M, Shi W, Teng IT, Creanga A, Jenkins C, Leung K, Liu T, Stancofski ED, Stephens T, Zhang B, Tsybovsky Y, Graham BS, Mascola JR, Sullivan NJ, Kwong PD
Science (2022) 376 pp. eabn8897-eabn8897 [ Pubmed: 35324257 DOI: doi:10.1126/science.abn8897 ]
- The light chain of sars-cov-2 antibody b1-182.1 (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- The light chain of sars-cov-2 antibody a19-61.1 (11 kDa, Protein from Homo sapiens)
- The heavy chain of sars-cov-2 antibody a19-61.1 (13 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike in complex with antibodies b1-182.1 and a19-61.1 (570 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (132 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- The heavy chain of sars-cov-2 antiboddy antibody b1-182.1 (13 kDa, Protein from Homo sapiens)
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 7l0l
Q-score: 0.555
Andrews SF, Raab JE, Gorman J, Gillespie RA, Cheung CSF, Rawi R, Cominsky LY, Boyington JC, Creanga A, Shen CH, Harris DR, Olia AS, Nazzari AF, Zhou T, Houser KV, Chen GL, Mascola JR, Graham BS, Kanekiyo M, Ledgerwood JE, Kwong PD, McDermott AB
Nat Med (2022) 28 pp. 373-382 [ DOI: doi:10.1038/s41591-021-01636-8 Pubmed: 35115707 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 316-310-1b11 heavy chain (25 kDa, Protein from Homo sapiens)
- Hemagglutinin ha1 chain (38 kDa, Protein from Influenza A virus (A/Canada/720/2005(H2N2)))
- Hemagglutinin ha2 chain (25 kDa, Protein from Influenza A virus (A/Canada/720/2005(H2N2)))
- 316-310-1b11 fab in complex with an h2 can05 ha trimer (Complex from Influenza A virus)
- 316-310-1b11 light chain (23 kDa, Protein from Homo sapiens)
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 7l2e
Q-score: 0.505
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819- [ DOI: doi:10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 4-18 heavy chain (24 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Prefusion sars-cov-2 spike glycoprotein in complex with 4-18 fab (Complex from Homo sapiens)
- 4-18 light chain (23 kDa, Protein from Homo sapiens)
Structure of SARS-CoV-2 spike at pH 4.5
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7jwy
Q-score: 0.612
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike at ph 4.5 (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
Structure of SARS-CoV-2 spike at pH 4.0
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 6xlu
Q-score: 0.63
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Consensus structure of SARS-CoV-2 spike at pH 5.5
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6xm0
Q-score: 0.583
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6xm3
Q-score: 0.562
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6xm4
Q-score: 0.559
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)