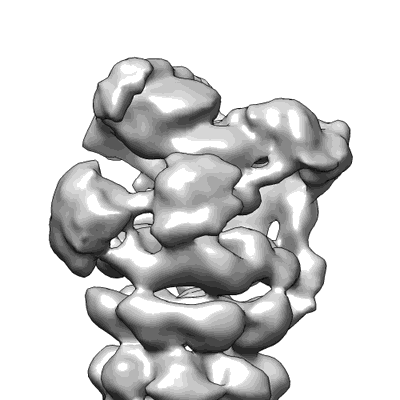
Structure of the yeast 26S proteasome lid sub-complex
Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC
eLife (2016) 5 pp. e13027-e13027 [ Pubmed: 26744777 DOI: doi:10.7554/elife.13027 ]
- Recombinant yeast 26s proteasome lid complex (370 kDa, Sample)
- 26s proteasome lid sub-complex (370 kDa, Protein from Saccharomyces cerevisiae)
Negative stain 3D reconstruction of the yeast 26S proteasome in complex with ubiquitin-bound Ubp6
Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A
Nat Struct Mol Biol (2015) 22 pp. 712-719 [ DOI: doi:10.1038/nsmb.3075 Pubmed: 26301997 ]
- Yeast 26s proteasome in complex with ubiquitin-bound ubp6 (1 MDa, Sample)
- Ubiquitin-bound ubp6 (65 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome (1 MDa, Protein from Saccharomyces cerevisiae)
Negative stain reconstruction of the Pex1/Pex6 complex in presence of ADP
Gardner BM, Chowdhury S, Lander GC, Martin A
J Mol Biol (2015) 427 pp. 1375-1388 [ Pubmed: 25659908 DOI: doi:10.1016/j.jmb.2015.01.019 ]
- Pex1/pex6 complex in presence of 3 mm adp (707 kDa, Sample)
- Peroxisomal atpase pex6 (115 kDa, Protein from Saccharomyces cerevisiae)
- Peroxisomal atpase pex1 (117 kDa, Protein from Saccharomyces cerevisiae)
Negative stain reconstruction of the Pex1/Pex6 complex in presence of ATP-gamma-S
Gardner BM, Chowdhury S, Lander GC, Martin A
J Mol Biol (2015) 427 pp. 1375-1388 [ Pubmed: 25659908 DOI: doi:10.1016/j.jmb.2015.01.019 ]
- Peroxisomal atpase pex6 (115 kDa, Protein from Saccharomyces cerevisiae)
- Pex1/pex6 complex in presence of 3 mm atp-gamma-s (707 kDa, Sample)
- Peroxisomal atpase pex1 (117 kDa, Protein from Saccharomyces cerevisiae)
Negative stain reconstruction of the Pex1/Pex6 complex in presence of ATP
Gardner BM, Chowdhury S, Lander GC, Martin A
J Mol Biol (2015) 427 pp. 1375-1388 [ Pubmed: 25659908 DOI: doi:10.1016/j.jmb.2015.01.019 ]
- Pex1/pex6 complex in presence of 3 mm atp (707 kDa, Sample)
- Peroxisomal atpase pex6 (115 kDa, Protein from Saccharomyces cerevisiae)
- Peroxisomal atpase pex1 (117 kDa, Protein from Saccharomyces cerevisiae)
Cryo EM density of microtubules stabilized by GMPCPP
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 3j6e
Q-score: 0.243
Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E
Cell (2014) 157 pp. 1117-1129 [ DOI: doi:10.1016/j.cell.2014.03.053 Pubmed: 24855948 ]
- Microtubule stabilized by gmpcpp (Sample)
- Kinesin (36 kDa, Protein from Homo sapiens)
- Beta tubulin (55 kDa, Protein from Sus scrofa)
- Alpha tubulin (55 kDa, Protein from Sus scrofa)
Cryo EM density of GDP-bound dynamic microtubules
Resolution: 4.9 Å
EM Method: Single-particle
Fitted PDBs: 3j6f
Q-score: 0.222
Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E
Cell (2014) 157 pp. 1117-1129 [ DOI: doi:10.1016/j.cell.2014.03.053 Pubmed: 24855948 ]
- Dynamic gdp-bound microtubule (Sample)
- Kinesin (36 kDa, Protein from Homo sapiens)
- Beta tubulin (55 kDa, Protein from Sus scrofa)
- Alpha tubulin (55 kDa, Protein from Sus scrofa)
Cryo EM density of microtubules stabilized by taxol
Resolution: 5.5 Å
EM Method: Single-particle
Fitted PDBs: 3j6g
Q-score: 0.194
Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E
Cell (2014) 157 pp. 1117-1129 [ DOI: doi:10.1016/j.cell.2014.03.053 Pubmed: 24855948 ]
- Microtubule stabilized by taxol (Sample)
- Kinesin (36 kDa, Protein from Homo sapiens)
- Beta tubulin (55 kDa, Protein from Sus scrofa)
- Alpha tubulin (55 kDa, Protein from Sus scrofa)