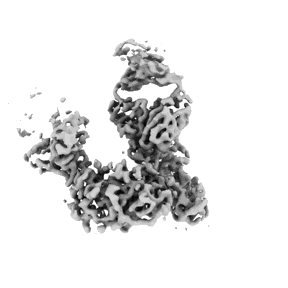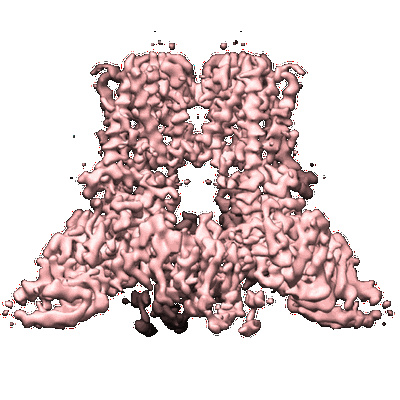Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Resolution: 3.48 Å
EM Method: Single-particle
Fitted PDBs: 8flj
Q-score: 0.35
Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin CC, Buyukyoruk M, Graham AB, Wilkinson RA, Triem L, Neselu K, Eng ET, Lander GC, Wiedenheft B
Nat Struct Mol Biol (2023) 30 pp. 1675-1685 [ Pubmed: 37710013 DOI: doi:10.1038/s41594-023-01097-2 ]
- Crispr leader, sense strand of dna (42 kDa, DNA from Pseudomonas aeruginosa PA14)
- Crispr-associated nuclease/helicase cas3 subtype i-f/ypest (121 kDa, Protein from Pseudomonas aeruginosa PA14)
- Integration host factor subunit alpha (11 kDa, Protein from Pseudomonas aeruginosa PA14)
- Ihf heterodimer (Complex from Pseudomonas aeruginosa PA14)
- Integration host factor subunit beta (10 kDa, Protein from Pseudomonas aeruginosa PA14)
- Crispr-associated endonuclease cas1 (38 kDa, Protein from Pseudomonas aeruginosa PA14)
- Crispr leader, repeat, and prespacer dna annealed to mimic a half-site integration intermediate. (Complex)
- Crispr repeat and prespacer, sense strand of dna (18 kDa, DNA from synthetic construct)
- Crispr leader and repeat, anti-sense strand of dna (52 kDa, DNA from Pseudomonas aeruginosa PA14)
- Cas1-cas2/3 heterohexameric integrase (Complex from Pseudomonas aeruginosa PA14)
- Cas1-cas2/3 integrase and ihf bound to crispr leader, repeat and foreign dna (122 kDa, Complex)
- Prespacer, anti-sense strand of dna (8 kDa, DNA from synthetic construct)
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8ffy
Q-score: 0.415
Kuhle B, Hirschi M, Doerfel LK, Lander GC, Schimmel P
Nat Commun (2023) 14 pp. 4794-4794 [ DOI: doi:10.1038/s41467-023-40354-2 Pubmed: 37558671 ]
- 5'-o-(n-(l-seryl)-sulfamoyl)adenosine (433 Da, Ligand)
- Serine--trna ligase, mitochondrial (58 kDa, Protein from Homo sapiens)
- Human mt-serrs in complex with mt-trna(uga-tl) and sersa (Complex from Homo sapiens)
- Mt-trna(uga-tl) (21 kDa, RNA from Homo sapiens)
Resolution: 3.83 Å
EM Method: Single-particle
Fitted PDBs: 7t6x
Q-score: 0.381
Torrents de la Pena A, Sliepen K, Eshun-Wilson L, Newby ML, Allen JD, Zon I, Koekkoek S, Chumbe A, Crispin M, Schinkel J, Lander GC, Sanders RW, Ward AB
Science (2022) 378 pp. 263-269 [ DOI: doi:10.1126/science.abn9884 Pubmed: 36264808 ]
- Igh505, lambda light chain (24 kDa, Protein from Homo sapiens)
- At12009 crossmab, light kappa chain (24 kDa, Protein from Homo sapiens)
- Ar4a fab, heavy chain (29 kDa, Protein from Homo sapiens)
- Ar4a, kappa light chain (25 kDa, Protein from Homo sapiens)
- Envelope glycoprotein e2 (40 kDa, Protein from Hepacivirus C)
- Igh505, heavy chain (52 kDa, Protein from Homo sapiens)
- Envelope glycoprotein e1 (20 kDa, Protein from Hepacivirus C)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- At12009 crossmab, heavy chain (29 kDa, Protein from Homo sapiens)
- Cryoem structure of full-length hepatitis c virus e1e2 glycoprotein in complex with ar4a, at12009 and igh505 fabs (Complex from Hepacivirus C)
Structure of the human TRPV3 channel in the apo conformation
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 6mho
Q-score: 0.446
Zubcevic L, Herzik Jr MA, Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, Lee SY
Nat Commun (2018) 9 pp. 4773-4773 [ Pubmed: 30429472 DOI: doi:10.1038/s41467-018-07117-w ]
Structure of the human TRPV3 channel in a putative sensitized conformation
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6mhs
Q-score: 0.392
Zubcevic L, Herzik Jr MA, Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, Lee SY
Nat Commun (2018) 9 pp. 4773-4773 [ Pubmed: 30429472 DOI: doi:10.1038/s41467-018-07117-w ]
Structure of human TRPV3 in the presence of 2-APB in C4 symmetry
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 6mhv
Q-score: 0.481
Zubcevic L, Herzik Jr MA, Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, Lee SY
Nat Commun (2018) 9 pp. 4773-4773 [ Pubmed: 30429472 DOI: doi:10.1038/s41467-018-07117-w ]
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1)
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6mhw
Q-score: 0.391
Zubcevic L, Herzik Jr MA, Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, Lee SY
Nat Commun (2018) 9 pp. 4773-4773 [ Pubmed: 30429472 DOI: doi:10.1038/s41467-018-07117-w ]
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (2)
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6mhx
Q-score: 0.372
Zubcevic L, Herzik Jr MA, Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, Lee SY
Nat Commun (2018) 9 pp. 4773-4773 [ Pubmed: 30429472 DOI: doi:10.1038/s41467-018-07117-w ]
Structure of the cold- and menthol-sensing ion channel TRPM8
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6bpq
Q-score: 0.395
Yin Y, Wu M, Zubcevic L, Borschel WF, Lander GC, Lee SY
Science (2018) 359 pp. 237-241 [ Pubmed: 29217583 DOI: doi:10.1126/science.aan4325 ]
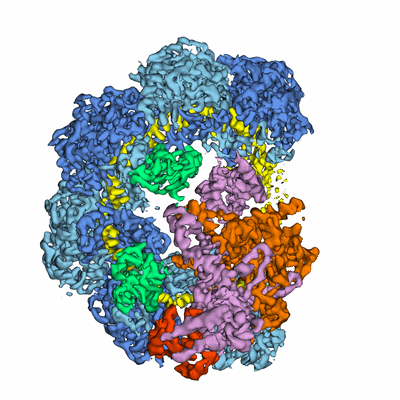
Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN, Hoffmann C, Nosaka L, Bondy-Denomy J, Maxwell KL, Davidson AR, Fischer ER, Lander GC, Wiedenheft B
Cell (2017) 169 pp. 47-57.e11 [ DOI: doi:10.1016/j.cell.2017.03.012 Pubmed: 28340349 ]
- Anti-crispr protein 30 (10 kDa, Protein from Pseudomonas phage D3112)
- Crispr-associated protein csy2 (36 kDa, Protein from Pseudomonas aeruginosa (strain UCBPP-PA14))
- Crispr rna (60-mer) (19 kDa, RNA from Pseudomonas aeruginosa UCBPP-PA14)
- Anti-crisprs acrf1 and acrf2 bound p. aeruginosa crrna-guided crispr surveillance(csy)complex. (450 kDa, Complex from Pseudomonas aeruginosa UCBPP-PA14)
- Crispr-associated protein csy3 (37 kDa, Protein from Pseudomonas aeruginosa (strain UCBPP-PA14))
- Crispr-associated protein csy1 (49 kDa, Protein from Pseudomonas aeruginosa (strain UCBPP-PA14))
- Crispr-associated endonuclease cas6/csy4 (21 kDa, Protein from Pseudomonas aeruginosa (strain UCBPP-PA14))
- Anti-crispr protein acr30-35 (8 kDa, Protein from Pseudomonas phage JBD30)