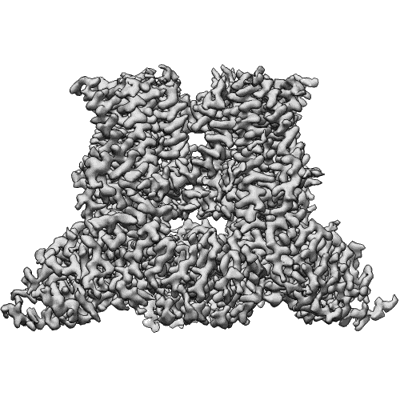
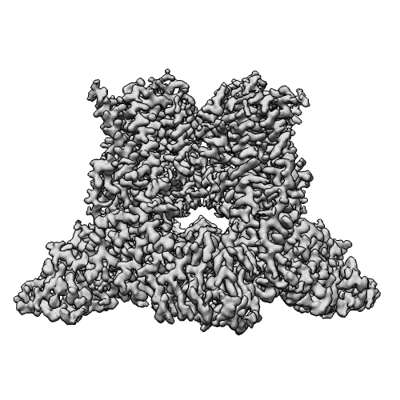
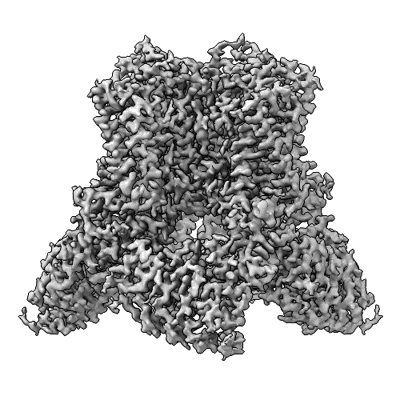Wildtype rat TRPV2 in nanodiscs bound to RR
Resolution: 3.47 Å
EM Method: Single-particle
Fitted PDBs: 8ffl
Q-score: 0.533
Pumroy RA, De Jesus-Perez JJ, Protopopova AD, Rocereta JA, Fluck EC, Fricke T, Lee BH, Rohacs T, Leffler A, Moiseenkova-Bell V
EMBO Rep (2024) 25 pp. 506-523 [ DOI: doi:10.1038/s44319-023-00050-0 Pubmed: 38225355 ]
Wildtype rat TRPV2 in nanodiscs bound to RR and 2-APB
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8ffm
Q-score: 0.606
Pumroy RA, De Jesus-Perez JJ, Protopopova AD, Rocereta JA, Fluck EC, Fricke T, Lee BH, Rohacs T, Leffler A, Moiseenkova-Bell V
EMBO Rep (2024) 25 pp. 506-523 [ DOI: doi:10.1038/s44319-023-00050-0 Pubmed: 38225355 ]
- Tetramer of wildtype rat trpv2 in nanodiscs bound to ruthenium red (340 kDa, Complex from Rattus norvegicus)
- 2-aminoethyl diphenylborinate (225 Da, Ligand)
- Transient receptor potential cation channel subfamily v member 2 (86 kDa, Protein from Rattus norvegicus)
- Ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) (559 Da, Ligand)
- 1,2-didecanoyl-sn-glycero-3-phosphoethanolamine (522 Da, Ligand)
RR-bound wildtype rabbit TRPV5 in nanodiscs
Resolution: 2.96 Å
EM Method: Single-particle
Fitted PDBs: 8ffn
Q-score: 0.577
Pumroy RA, De Jesus-Perez JJ, Protopopova AD, Rocereta JA, Fluck EC, Fricke T, Lee BH, Rohacs T, Leffler A, Moiseenkova-Bell V
EMBO Rep (2024) 25 pp. 506-523 [ DOI: doi:10.1038/s44319-023-00050-0 Pubmed: 38225355 ]
- (2s)-3-(hexadecanoyloxy)-2-[(9z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate (760 Da, Ligand)
- Transient receptor potential cation channel subfamily v member 5 (83 kDa, Protein from Oryctolagus cuniculus)
- Tetramer of wildtype rabbit trpv5 in nanodiscs bound to ruthenium red (335 kDa, Complex from Oryctolagus cuniculus)
- Ergosterol (396 Da, Ligand)
- Ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) (559 Da, Ligand)
Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2 and ruthenium red
Resolution: 2.65 Å
EM Method: Single-particle
Fitted PDBs: 8ffq
Q-score: 0.624
Pumroy RA, De Jesus-Perez JJ, Protopopova AD, Rocereta JA, Fluck EC, Fricke T, Lee BH, Rohacs T, Leffler A, Moiseenkova-Bell V
EMBO Rep (2024) 25 pp. 506-523 [ DOI: doi:10.1038/s44319-023-00050-0 Pubmed: 38225355 ]
- Tetramer of wildtype rabbit trpv5 into nanodiscs in complex with ruthenium red and pi(4,5)p2 (335 kDa, Complex from Oryctolagus cuniculus)
- Transient receptor potential cation channel subfamily v member 5 (83 kDa, Protein from Oryctolagus cuniculus)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- Ergosterol (396 Da, Ligand)
- Ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) (559 Da, Ligand)
Cryo-EM structure of wildtype rabbit TRPV5 with PI(4,5)P2 in nanodiscs
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8ffo
Q-score: 0.498
Lee BH, De Jesus Perez JJ, Moiseenkova-Bell V, Rohacs T
Nat Commun (2023) 14 pp. 5883-5883 [ DOI: doi:10.1038/s41467-023-41577-z Pubmed: 37735536 ]
- Tetramer of wildtype rabbit trpv5 in nanodiscs in complex with pi(4,5)p2 (335 kDa, Complex from Oryctolagus cuniculus)
- Transient receptor potential cation channel subfamily v member 5 (83 kDa, Protein from Oryctolagus cuniculus)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- Ergosterol (396 Da, Ligand)
- [(2r)-2-octanoyloxy-3-[oxidanyl-[(1r,2r,3s,4r,5r,6s)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate (746 Da, Ligand)
Wildtype rabbit TRPV5 in nanodiscs in the presence of oleoyl coenzyme A, Closed stated
Resolution: 3.09 Å
EM Method: Single-particle
Fitted PDBs: 8fhh
Q-score: 0.554
Lee BH, De Jesus Perez JJ, Moiseenkova-Bell V, Rohacs T
Nat Commun (2023) 14 pp. 5883-5883 [ DOI: doi:10.1038/s41467-023-41577-z Pubmed: 37735536 ]
- Tetramer of wildtype rabbit trpv5 in nanodiscs in the presence of oleoyl coenzyme a (335 kDa, Complex from Oryctolagus cuniculus)
- Transient receptor potential cation channel subfamily v member 5 (83 kDa, Protein from Oryctolagus cuniculus)
- (2s)-3-(hexadecanoyloxy)-2-[(9z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate (760 Da, Ligand)
- Ergosterol (396 Da, Ligand)
Wildtype rabbit TRPV5 in nanodiscs in complex with oleoyl coenzyme A, Open stated
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 8fhi
Q-score: 0.504
Lee BH, De Jesus Perez JJ, Moiseenkova-Bell V, Rohacs T
Nat Commun (2023) 14 pp. 5883-5883 [ DOI: doi:10.1038/s41467-023-41577-z Pubmed: 37735536 ]
- Tetramer of wildtype rabbit trpv5 in nanodiscs in complex with oleoyl coenzyme a (335 kDa, Complex from Oryctolagus cuniculus)
- (2s)-3-(hexadecanoyloxy)-2-[(9z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate (760 Da, Ligand)
- S-{(3r,5r,9r)-1-[(2r,3s,4r,5r)-5-(6-amino-9h-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9z)-octadec-9-enethioate (non-preferred name) (1 kDa, Ligand)
- Transient receptor potential cation channel subfamily v member 5 (83 kDa, Protein from Oryctolagus cuniculus)
- Ergosterol (396 Da, Ligand)
26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS)
Resolution: 7.0 Å
EM Method: Single-particle
Fitted PDBs: 7qo4
Q-score: 0.112
Hung KYS, Klumpe S, Eisele MR, Elsasser S, Tian G, Sun S, Moroco JA, Cheng TC, Joshi T, Seibel T, Van Dalen D, Feng XH, Lu Y, Ovaa H, Engen JR, Lee BH, Rudack T, Sakata E, Finley D
Nat Commun (2022) 13 pp. 838- [ DOI: doi:10.1038/s41467-022-28186-y ]
- Rpt3 isoform 1 (47 kDa, Protein from baker's yeast)
- Ubp6-ubvs (Complex from Saccharomyces cerevisiae)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Polyubiquitin-b (8 kDa, Protein from Homo sapiens)
- Yeast wt 26s proteasome body2 (atpase, rpn1) (Complex from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- 26s proteasome wt-ubp6-ubvs complex in the si state (2 MDa, Complex)
- Rpt2 isoform 1 (48 kDa, Protein from baker's yeast)
- Ubiquitin carboxyl-terminal hydrolase (57 kDa, Protein from Saccharomyces cerevisiae)
- Rpt1 isoform 1 (52 kDa, Protein from baker's yeast)
- Rpt4 isoform 1 (49 kDa, Protein from baker's yeast)
- Rpt5 isoform 1 (48 kDa, Protein from baker's yeast)
- Rpt6 isoform 1 (45 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn1 (109 kDa, Protein from baker's yeast)
Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid)
Resolution: 6.1 Å
EM Method: Single-particle
Fitted PDBs: 7qo3
Q-score: 0.133
Hung KYS, Klumpe S, Eisele MR, Elsasser S, Tian G, Sun S, Moroco JA, Cheng TC, Joshi T, Seibel T, Van Dalen D, Feng XH, Lu Y, Ovaa H, Engen JR, Lee BH, Rudack T, Sakata E, Finley D
Nat Commun (2022) 13 pp. 838-838 [ DOI: doi:10.1038/s41467-022-28186-y Pubmed: 35149681 ]
- 26s proteasome-ubp6 complex (si state) (Complex from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-3 (28 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-4 (28 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn13 (17 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-6 (25 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn5 (51 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn9 (45 kDa, Protein from Saccharomyces cerevisiae)
- Probable proteasome subunit alpha type-7 (31 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-6 (26 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-7 (29 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-5 (28 kDa, Protein from Saccharomyces cerevisiae)
- Ubiquitin carboxyl-terminal hydrolase rpn11 (34 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-4 (22 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn8 (38 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn12 (31 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-1 (23 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-2 (27 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn1 (109 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-5 (31 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome complex subunit sem1 (10 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit alpha type-1 (28 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn10 (29 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn3 (60 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn7 (49 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn6 (49 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-2 (28 kDa, Protein from Saccharomyces cerevisiae)
- Proteasome subunit beta type-3 (22 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit rpn2 (104 kDa, Protein from Saccharomyces cerevisiae)
26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state
Resolution: 6.0 Å
EM Method: Single-particle
Fitted PDBs: 7qo5
Q-score: 0.133
Hung KYS, Klumpe S, Eisele MR, Elsasser S, Tian G, Sun S, Moroco JA, Cheng TC, Joshi T, Seibel T, Van Dalen D, Feng XH, Lu Y, Ovaa H, Engen JR, Lee BH, Rudack T, Sakata E, Finley D
Nat Commun (2022) 13 pp. 838-838 [ DOI: doi:10.1038/s41467-022-28186-y Pubmed: 35149681 ]
- Proteasome subunit alpha type-2 (27 kDa, Protein from baker's yeast)
- Proteasome subunit alpha type-6 (25 kDa, Protein from baker's yeast)
- Proteasome subunit alpha type-1 (28 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit 7 homolog (52 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit 8 homolog (45 kDa, Protein from baker's yeast)
- Polyubiquitin-b (8 kDa, Protein from Homo sapiens)
- 26s proteasome regulatory subunit 4 homolog (48 kDa, Protein from baker's yeast)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- 26s proteasome regulatory subunit rpn5 (51 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn12 (31 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn1 (109 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn7 (49 kDa, Protein from baker's yeast)
- Magnesium ion (24 Da, Ligand)
- Yeast rpt1-rk 26s proteasome in si state (Complex from Saccharomyces cerevisiae)
- Proteasome subunit beta type-2 (28 kDa, Protein from baker's yeast)
- Proteasome subunit beta type-3 (22 kDa, Protein from baker's yeast)
- 26s proteasome rpt1-rk -ubp6-ubvs complex in the si state (Complex)
- Ubiquitin carboxyl-terminal hydrolase 6 (57 kDa, Protein from Saccharomyces cerevisiae)
- 26s proteasome regulatory subunit 6a (48 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn10 (29 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn6 (49 kDa, Protein from baker's yeast)
- Proteasome subunit alpha type-5 (28 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn13 (17 kDa, Protein from baker's yeast)
- Ubp6-ubvs complex (Complex from Saccharomyces cerevisiae)
- Proteasome subunit beta type-4 (22 kDa, Protein from baker's yeast)
- Ubiquitin carboxyl-terminal hydrolase rpn11 (34 kDa, Protein from baker's yeast)
- 26s proteasome subunit rpt4 (49 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn2 (104 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn9 (45 kDa, Protein from baker's yeast)
- 26s proteasome complex subunit sem1 (10 kDa, Protein from baker's yeast)
- Proteasome subunit beta type-5 (31 kDa, Protein from baker's yeast)
- Proteasome subunit beta type-7 (29 kDa, Protein from baker's yeast)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- 26s proteasome regulatory subunit 6b homolog (47 kDa, Protein from baker's yeast)
- Probable proteasome subunit alpha type-7 (31 kDa, Protein from baker's yeast)
- Proteasome subunit beta type-6 (26 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn3 (60 kDa, Protein from baker's yeast)
- Proteasome subunit beta type-1 (23 kDa, Protein from baker's yeast)
- 26s proteasome regulatory subunit rpn8 (38 kDa, Protein from baker's yeast)
- Proteasome subunit alpha type-4 (28 kDa, Protein from baker's yeast)
- Proteasome subunit alpha type-3 (28 kDa, Protein from baker's yeast)