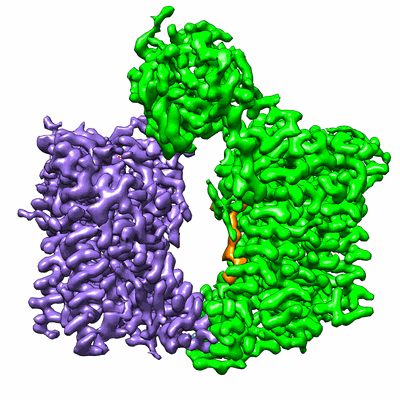
Structure of the S. cerevisiae ER membrane complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 6wb9
Q-score: 0.532
Bai L, You Q, Feng X, Kovach A, Li H
Nature (2020) 584 pp. 475-478 [ Pubmed: 32494008 DOI: doi:10.1038/s41586-020-2389-3 ]
- Er membrane protein complex subunit 3 (28 kDa, Protein from Saccharomyces cerevisiae W303)
- Er membrane protein complex subunit 4 (21 kDa, Protein from Saccharomyces cerevisiae W303)
- Er membrane protein complex subunit 5 (15 kDa, Protein from Saccharomyces cerevisiae W303)
- Protein sop4 (26 kDa, Protein from Saccharomyces cerevisiae W303)
- Er membrane protein complex subunit 1 (87 kDa, Protein from Saccharomyces cerevisiae W303)
- Er membrane complex (Complex from Saccharomyces cerevisiae W303)
- (2s)-3-(hexadecanoyloxy)-2-[(9z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate (760 Da, Ligand)
- Er membrane protein complex subunit 6 (12 kDa, Protein from Saccharomyces cerevisiae W303)
- Endoplasmic reticulum membrane protein complex subunit 10 (22 kDa, Protein from Saccharomyces cerevisiae W303)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Er membrane protein complex subunit 2 (33 kDa, Protein from Saccharomyces cerevisiae W303)
Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6psx
Q-score: 0.383
Bai L, Kovach A, You Q, Hsu HC, Zhao G, Li H
Nat Commun (2019) 10 pp. 4142-4142 [ DOI: doi:10.1038/s41467-019-12191-9 Pubmed: 31515475 ]
- Drs2p-cdc50p complex (Complex from Saccharomyces cerevisiae W303)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cell division control protein 50 (45 kDa, Protein from Saccharomyces cerevisiae W303)
- Probable phospholipid-transporting atpase drs2 (153 kDa, Protein from Saccharomyces cerevisiae W303)
Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 6psy
Q-score: 0.506
Bai L, Kovach A, You Q, Hsu HC, Zhao G, Li H
Nat Commun (2019) 10 pp. 4142-4142 [ DOI: doi:10.1038/s41467-019-12191-9 Pubmed: 31515475 ]
- Drs2p-cdc50p complex (Complex from Saccharomyces cerevisiae W303)
- Probable phospholipid-transporting atpase drs2 (153 kDa, Protein from Saccharomyces cerevisiae W303)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cell division control protein 50 (45 kDa, Protein from Saccharomyces cerevisiae W303)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6p25
Q-score: 0.47
Bai L, Kovach A, You Q, Kenny A, Li H
Nat Struct Mol Biol (2019) 26 pp. 704-711 [ Pubmed: 31285605 DOI: doi:10.1038/s41594-019-0262-6 ]
- (3r)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate (644 Da, Ligand)
- Acceptor peptide (478 Da, Protein from Saccharomyces cerevisiae W303)
- Protein o-mannosyl transferase complex (Complex from Saccharomyces cerevisiae W303)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dolichyl-phosphate-mannose--protein mannosyltransferase 2 (86 kDa, Protein from Saccharomyces cerevisiae W303)
- Dolichyl-phosphate-mannose--protein mannosyltransferase 1 (92 kDa, Protein from Saccharomyces cerevisiae W303)
Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6p2r
Q-score: 0.454
Bai L, Kovach A, You Q, Kenny A, Li H
Nat Struct Mol Biol (2019) 26 pp. 704-711 [ Pubmed: 31285605 DOI: doi:10.1038/s41594-019-0262-6 ]
- (3r)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate (644 Da, Ligand)
- Protein o-mannosyl transferase complex (Complex from Saccharomyces cerevisiae W303)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dolichyl-phosphate-mannose--protein mannosyltransferase 2 (86 kDa, Protein from Saccharomyces cerevisiae W303)
- Dolichyl-phosphate-mannose--protein mannosyltransferase 1 (92 kDa, Protein from Saccharomyces cerevisiae W303)
The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 6c26
Q-score: 0.503
Bai L, Wang T, Zhao G, Kovach A, Li H
Nature (2018) 555 pp. 328-333 [ DOI: doi:10.1038/nature25755 Pubmed: 29466327 ]
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit ost5 (9 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit ost4 (3 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3 (39 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Oligosaccharyl transferase complex (Complex from Saccharomyces cerevisiae)
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit ost2 (14 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit swp1 (31 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit stt3 (81 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit wbp1 (49 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- (4r,7r)-4-hydroxy-n,n,n-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium (636 Da, Ligand)
- Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 (54 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))