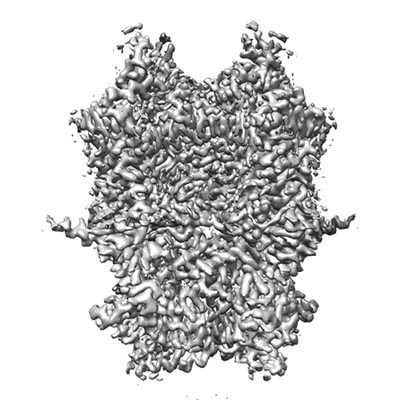
Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 8yhd
Q-score: 0.5
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- A protein (Complex from metagenome)
- Zinc ion (65 Da, Ligand)
- A protein (27 kDa, Protein from metagenome)
- Rna (53-mer) (21 kDa, RNA from metagenome)
- A protein (22 kDa, Protein from metagenome)
- A protein (70 kDa, Protein from metagenome)
- Rna (35-mer) (16 kDa, RNA from metagenome)
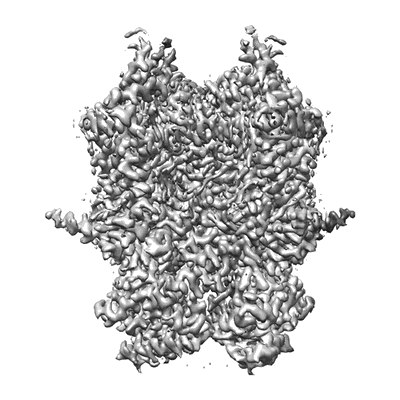
Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 8yhe
Q-score: 0.506
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- Zinc ion (65 Da, Ligand)
- Rna (46-mer) (15 kDa, RNA from metagenome)
- Rna (29-mer) (16 kDa, RNA from metagenome)
- Protein structure (70 kDa, Protein from metagenome)
- Protein structure (27 kDa, Protein from metagenome)
- Protein structure (22 kDa, Protein from metagenome)
- Protein structure (Complex from metagenome)
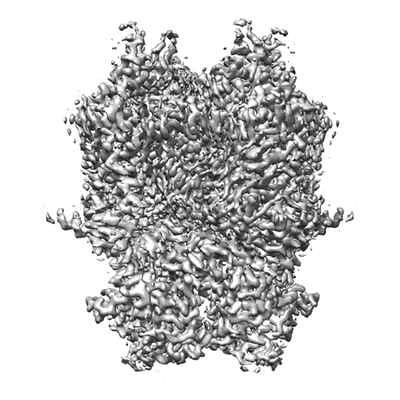
Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 8z4j
Q-score: 0.507
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- A protein (Complex from metagenome)
- Zinc ion (65 Da, Ligand)
- Rna (34-mer) (19 kDa, RNA from metagenome)
- Protein structure (27 kDa, Protein from metagenome)
- Protein structure (22 kDa, Protein from metagenome)
- Rna (38-mer) (19 kDa, RNA from metagenome)
- Protein structure (70 kDa, Protein from metagenome)
Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8z4l
Q-score: 0.531
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- A protein (Complex from metagenome)
- Zinc ion (65 Da, Ligand)
- A protein (27 kDa, Protein from metagenome)
- Rna (49-mer) (19 kDa, RNA from metagenome)
- A protein (22 kDa, Protein from metagenome)
- A protein (70 kDa, Protein from metagenome)
- Rna (40-mer) (19 kDa, RNA from metagenome)
Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8z99
Q-score: 0.487
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- A protein (Complex from metagenome)
- Zinc ion (65 Da, Ligand)
- A protein (27 kDa, Protein from metagenome)
- A protein (22 kDa, Protein from metagenome)
- A protein (70 kDa, Protein from metagenome)
- Rna (49-mer) (19 kDa, RNA from metagenome)
- Rna (54-mer) (19 kDa, RNA from metagenome)
Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 8z9c
Q-score: 0.528
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- Rna (48-mer) (19 kDa, RNA from metagenome)
- Zinc ion (65 Da, Ligand)
- Protein structure (27 kDa, Protein from metagenome)
- Protein structure (22 kDa, Protein from metagenome)
- Protein structure (70 kDa, Protein from metagenome)
- Rna (41-mer) (17 kDa, RNA from metagenome)
- Protein structure (Complex from metagenome)
Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 8z9e
Q-score: 0.496
Yang J, Li X, He Q, Wang X, Tang J, Wang T, Zhang Y, Yu F, Zhang S, Liu Z, Zhang L, Liao F, Yin H, Zhao H, Deng Z, Zhang H
Nature (2024) 633 pp. 465-472 [ Pubmed: 39143216 DOI: doi:10.1038/s41586-024-07815-0 ]
- Zinc ion (65 Da, Ligand)
- Rna (34-mer) (19 kDa, RNA from metagenome)
- Protein structure (27 kDa, Protein from metagenome)
- Rna (39-mer) (19 kDa, RNA from metagenome)
- Protein structure (22 kDa, Protein from metagenome)
- Protein structure (70 kDa, Protein from metagenome)
- Protein structure (Complex from metagenome)
Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 6wej
Q-score: 0.582
Zheng X, Fu Z, Su D, Zhang Y, Li M, Pan Y, Li H, Li S, Grassucci RA, Ren Z, Hu Z, Li X, Zhou M, Li G, Frank J, Yang J
Nat Struct Mol Biol (2020) 27 pp. 625-634 [ DOI: doi:10.1038/s41594-020-0433-5 Pubmed: 32483338 ]
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- Structure of cgmp-unbound wt tax-4 reconstituted in lipid nanodiscs (Complex from Caenorhabditis elegans)
- Cyclic nucleotide-gated cation channel (83 kDa, Protein from Caenorhabditis elegans)
Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6wek
Q-score: 0.593
Zheng X, Fu Z, Su D, Zhang Y, Li M, Pan Y, Li H, Li S, Grassucci RA, Ren Z, Hu Z, Li X, Zhou M, Li G, Frank J, Yang J
Nat Struct Mol Biol (2020) 27 pp. 625-634 [ DOI: doi:10.1038/s41594-020-0433-5 Pubmed: 32483338 ]
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- Cyclic guanosine monophosphate (345 Da, Ligand)
- Cyclic nucleotide-gated cation channel (83 kDa, Protein from Caenorhabditis elegans)
- Structure of cgmp-bound wt tax-4 reconstituted in lipid nanodiscs (Complex from Caenorhabditis elegans)
Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 6wel
Q-score: 0.616
Zheng X, Fu Z, Su D, Zhang Y, Li M, Pan Y, Li H, Li S, Grassucci RA, Ren Z, Hu Z, Li X, Zhou M, Li G, Frank J, Yang J
Nat Struct Mol Biol (2020) 27 pp. 625-634 [ DOI: doi:10.1038/s41594-020-0433-5 Pubmed: 32483338 ]
- 1,2-dilauroyl-sn-glycero-3-phosphate (535 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (758 Da, Ligand)
- Structure of cgmp-unbound f403v/v407a mutant tax-4 reconstituted in lipid nanodiscs (Complex from Caenorhabditis elegans)
- Cyclic nucleotide-gated cation channel (83 kDa, Protein from Caenorhabditis elegans)