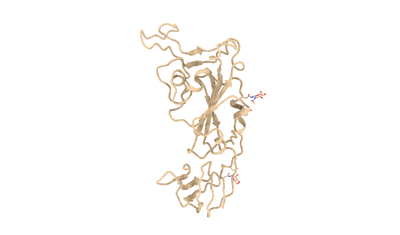
SARS-CoV-2 Omicron BA.3 variant spike (local)
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 7xiz
Q-score: 0.392
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
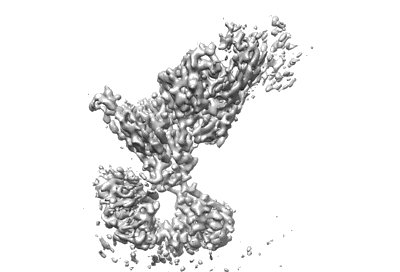
Local structure of BD55-1239 Fab and SARS-COV2 Omicron RBD complex
Resolution: 3.51 Å
EM Method: Single-particle
Fitted PDBs: 7wrl
Q-score: 0.472
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
- Bd55-1239h (13 kDa, Protein from SARS coronavirus B012)
- Bd55-1239l (11 kDa, Protein from SARS coronavirus B012)
- Spike protein s1 (21 kDa, Protein from SARS coronavirus B012)
- Local structure of bd55-1239 fab and sars-cov2 omicron rbd complex (Complex from SARS coronavirus B012)
Local structure of BD55-3372 and delta spike
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7wro
Q-score: 0.482
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
- Delta rbd (Complex from Severe acute respiratory syndrome coronavirus)
- 3372l (11 kDa, Protein from Homo sapiens)
- Local structure of bd55-3372 and delta rbd (Complex)
- 3372h (13 kDa, Protein from Homo sapiens)
- Bd55-3372 (Complex from Homo sapiens)
- Spike protein s1 (20 kDa, Protein from Severe acute respiratory syndrome coronavirus)
Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD
Resolution: 3.26 Å
EM Method: Single-particle
Fitted PDBs: 7wrz
Q-score: 0.544
Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS
Nature (2022) 608 pp. 593-602 [ DOI: doi:10.1038/s41586-022-04980-y Pubmed: 35714668 ]
- Bd55-5840l (11 kDa, Protein from SARS coronavirus B012)
- Spike protein s1 (21 kDa, Protein from SARS coronavirus B012)
- Local resolution of bd55-5840 fab and sars-cov2 omicron rbd (Complex from Homo sapiens)
- Bd55-5840h (13 kDa, Protein from SARS coronavirus B012)
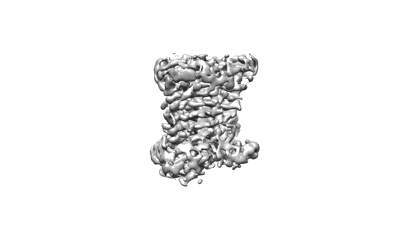
Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with PAMP-12, local
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7vv0
Q-score: 0.469
Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP
Nature (2021) 600 pp. 164-169 [ DOI: doi:10.1038/s41586-021-04077-y Pubmed: 34789875 ]
- Peptide from pro-adrenomedullin (1 kDa, Protein from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- Cryo-em structure of pseudoallergen receptor mrgprx2 complex with pamp-12, local (Complex from Homo sapiens)
- Mas-related g-protein coupled receptor member x2 (37 kDa, Protein from Homo sapiens)
Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with linear cortistatin-14, local
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 7vv4
Q-score: 0.471
Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP
Nature (2021) 600 pp. 164-169 [ DOI: doi:10.1038/s41586-021-04077-y Pubmed: 34789875 ]
- Cryo-em structure of pseudoallergen receptor mrgprx2 complex with linear cortistatin-14, local (Complex from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- Circular cortistatin-14 (1 kDa, Protein from Homo sapiens)
- Mas-related g-protein coupled receptor member x2 (37 kDa, Protein from Homo sapiens)
Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local)
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7vv6
Q-score: 0.502
Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP
Nature (2021) 600 pp. 164-169 [ DOI: doi:10.1038/s41586-021-04077-y Pubmed: 34789875 ]
- 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{n}-methyl-ethanamine (519 Da, Ligand)
- Cryo-em structure of pseudoallergen receptor mrgprx2 complex with c48/80 (local) (Complex from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- Mas-related g-protein coupled receptor member x2 (37 kDa, Protein from Homo sapiens)
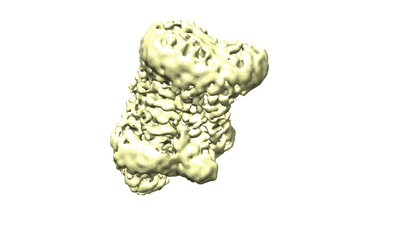
Cryo-EM structure of the Slo2.2 Na+-activated K+ channel
Resolution: 5.2 Å
EM Method: Single-particle
Fitted PDBs: 5a6g
Q-score: 0.224
Hite RK, Yuan P, Li Z, Hsuing Y, Walz T, MacKinnon R
Nature (2015) 527 pp. 198-203 [ Pubmed: 26436452 DOI: doi:10.1038/NATURE14958 ]
- Slo2.2 (550 kDa, Protein from Gallus gallus)
- Chicken slo2.2 (550 kDa, Sample)