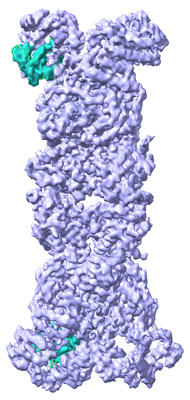
Tp-PSI-FCPI-S in Thalassiosira pseudonana
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8zet
Q-score: 0.485
Feng Y, Li Z, Yang Y, Shen L, Li X, Liu X, Zhang X, Zhang J, Ren F, Wang Y, Liu C, Han G, Wang X, Kuang T, Shen JR, Wang W
J Integr Plant Biol (2024) [ DOI: doi:10.1111/jipb.13816 Pubmed: 39670505 ]
- Photosystem i reaction center subunit xii (3 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i reaction center subunit ix (4 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Phylloquinone (450 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Photosystem i reaction center subunit viii (3 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i p700 chlorophyll a apoprotein a2 (82 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Beta-carotene (536 Da, Ligand)
- Chlorophyll c1 (610 Da, Ligand)
- Photosystem i reaction center subunit ii (14 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i reaction center subunit iv (7 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Fucoxanthin chl a/c light-harvesting protein, major type (18 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Pt17531-like protein (18 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Tp-redcap (20 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- (3s,3's,5r,5'r,6s,6'r,8'r)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate (658 Da, Ligand)
- Fucoxanthin chlorophyll a/c-binding protein lhcq8 (18 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i iron-sulfur center (8 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i p700 chlorophyll a apoprotein a1 (82 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- (3s,3'r,5r,6s,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol (582 Da, Ligand)
- Tp-psar (9 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- (1~{r})-3,5,5-trimethyl-4-[(1~{e},3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e})-3,7,12,16-tetramethyl-18-[(4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol (566 Da, Ligand)
- 1,2-di-o-acyl-3-o-[6-deoxy-6-sulfo-alpha-d-glucopyranosyl]-sn-glycerol (795 Da, Ligand)
- Photosystem i reaction center subunit iii (17 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Photosystem i reaction center subunit psa29 (13 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Fucoxanthin chl a/c light-harvesting protein (18 kDa, Protein from Thalassiosira pseudonana CCMP1335)
- Digalactosyl diacyl glycerol (dgdg) (949 Da, Ligand)
- Tp-psi-fcpi-s (Complex from Thalassiosira pseudonana CCMP1335)
- Iron/sulfur cluster (351 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- Photosystem i reaction center subunit xi (15 kDa, Protein from Thalassiosira pseudonana CCMP1335)
Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2
Resolution: 3.16 Å
EM Method: Single-particle
Fitted PDBs: 8z8j
Q-score: 0.515
Xue L, Chang T, Li Z, Wang C, Zhao H, Li M, Tang P, Wen X, Yu M, Wu J, Bao X, Wang X, Gong P, He J, Chen X, Xiong X
Nat Commun (2024) 15 pp. 4620-4620 [ DOI: doi:10.1038/s41467-024-48848-3 Pubmed: 38816392 ]
- Rna-directed rna polymerase catalytic subunit (81 kDa, Protein from Thogoto virus (isolate SiAr 126))
- Polymerase acidic protein (71 kDa, Protein from Thogoto virus (isolate SiAr 126))
- Rna (5'-r(*ap*gp*ap*gp*ap*ap*ap*up*cp*ap*ap*gp*gp*cp*ap*gp*up*u)-3') (5 kDa, RNA from Thogoto virus (isolate SiAr 126))
- Cryo-em structure of thogoto virus polymerase in transcription pre-initiation conformation 2 (Complex from Thogoto virus (isolate SiAr 126))
- Rna (5'-r(*gp*ap*cp*up*gp*cp*cp*up*gp*up*up*up*up*up*gp*cp*u)-3') (5 kDa, RNA from Thogoto virus (isolate SiAr 126))
- Polymerase basic protein 2 (94 kDa, Protein from Thogoto virus (isolate SiAr 126))
Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation
Resolution: 3.06 Å
EM Method: Single-particle
Fitted PDBs: 8z8x
Q-score: 0.525
Xue L, Chang T, Li Z, Wang C, Zhao H, Li M, Tang P, Wen X, Yu M, Wu J, Bao X, Wang X, Gong P, He J, Chen X, Xiong X
Nat Commun (2024) 15 pp. 4620-4620 [ DOI: doi:10.1038/s41467-024-48848-3 Pubmed: 38816392 ]
- Phosphomethylphosphonic acid guanylate ester (521 Da, Ligand)
- Rna-directed rna polymerase catalytic subunit (81 kDa, Protein from Thogoto virus (isolate SiAr 126))
- Polymerase acidic protein (71 kDa, Protein from Thogoto virus (isolate SiAr 126))
- Rna (5'-r(*ap*gp*ap*gp*ap*ap*ap*up*cp*ap*ap*gp*gp*cp*ap*gp*up*u)-3') (5 kDa, RNA from Thogoto virus (isolate SiAr 126))
- Rna (5'-r(*gp*ap*cp*up*gp*cp*cp*up*gp*up*up*up*up*up*gp*cp*u)-3') (5 kDa, RNA from Thogoto virus (isolate SiAr 126))
- Polymerase basic protein 2 (94 kDa, Protein from Thogoto virus (isolate SiAr 126))
- Cryo-em structure of thogoto virus polymerase in a transcription initiation conformation (Complex from Thogoto virus (isolate SiAr 126))
Cryo-EM structure of tail tube protein
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 8xkn
Q-score: 0.542
Yin H, Li X, Wang X, Zhang C, Gao J, Yu G, He Q, Yang J, Liu X, Wei Y, Li Z, Zhang H
Nat Commun (2024) 15 pp. 2692-2692 [ DOI: doi:10.1038/s41467-024-47030-z Pubmed: 38538592 ]
- Cryo-em structure of tail tube protein (Complex from Bacillus halotolerans)
- A protein (29 kDa, Protein from Bacillus halotolerans)
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 8i3s
Q-score: 0.4
Liu Q, Zhao H, Li Z, Zhang Z, Huang R, Gu M, Zhuang K, Xiong Q, Chen X, Yu W, Qian S, Zhang Y, Tan X, Zhang M, Yu F, Guo M, Huang Z, Wang X, Xiang W, Wu B, Mei F, Cai K, Zhou L, Zhou L, Wu Y, Yan H, Cao S, Lan K, Chen Y
Signal Transduct Target Ther (2023) 8 pp. 347-347 [ Pubmed: 37704615 DOI: doi:10.1038/s41392-023-01615-0 ]
- Light chain of fab 7b3 (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Heavy chain od fab 7b3 (24 kDa, Protein from Homo sapiens)
- Spike-fab complex (Complex from Homo sapiens)
- Spike glycoprotein (137 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8i3u
Q-score: 0.462
Liu Q, Zhao H, Li Z, Zhang Z, Huang R, Gu M, Zhuang K, Xiong Q, Chen X, Yu W, Qian S, Zhang Y, Tan X, Zhang M, Yu F, Guo M, Huang Z, Wang X, Xiang W, Wu B, Mei F, Cai K, Zhou L, Zhou L, Wu Y, Yan H, Cao S, Lan K, Chen Y
Signal Transduct Target Ther (2023) 8 pp. 347-347 [ Pubmed: 37704615 DOI: doi:10.1038/s41392-023-01615-0 ]
- Heavy chain of fab 14b1 (16 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike-fab complex (Complex from Severe acute respiratory syndrome coronavirus 2)
- Light chain of fab 14b1 (14 kDa, Protein from Homo sapiens)
- Spike glycoprotein (137 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of the NmeCas9-sgRNA-AcrIIC4 ternary complex
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 8ja0
Q-score: 0.442
Li X, Liao F, Gao J, Song G, Zhang C, Ji N, Wang X, Wen J, He J, Wei Y, Zhang H, Li Z, Yu G, Yin H
Nucleic Acids Res (2023) 51 pp. 9442-9451 [ DOI: doi:10.1093/nar/gkad669 Pubmed: 37587688 ]
- Crispr-associated endonuclease cas9 (124 kDa, Protein from Neisseria meningitidis)
- Uncharacterized protein (9 kDa, Protein from Haemophilus parainfluenzae)
- Rna (117-mer) (37 kDa, RNA from Neisseria meningitidis)
- A protein complex (Complex from Neisseria meningitidis)