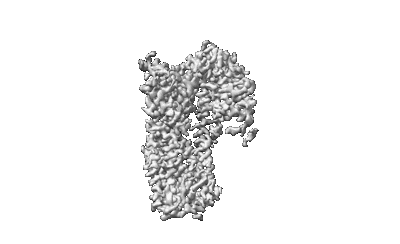
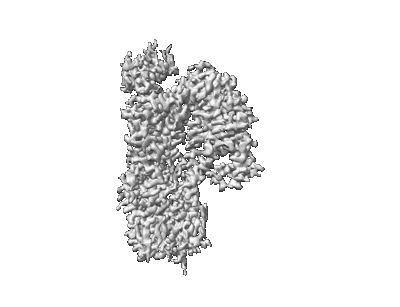
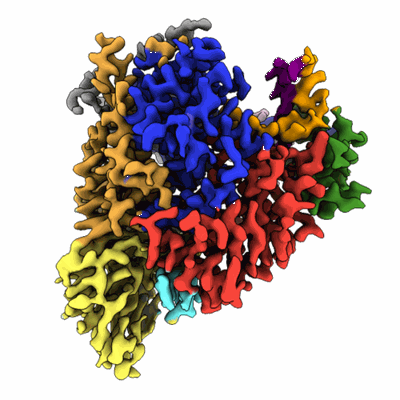
Cryo-EM structure of human receptor with G proteins
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8ikg
Q-score: 0.432
Shen S, Wu C, Lin G, Yang X, Zhou Y, Zhao C, Miao Z, Tian X, Wang K, Yang Z, Liu Z, Guo N, Li Y, Xia A, Zhou P, Liu J, Yan W, Ke B, Yang S, Shao Z
PNAS (2024) 121 pp. e2321532121-e2321532121 [ Pubmed: 38830102 DOI: doi:10.1073/pnas.2321532121 ]
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Scfv16 (28 kDa, Protein from Mus musculus)
- Cannabinoid receptor 1 (35 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Gi heterotrimer (Complex from Homo sapiens)
- Cannabinoid receptor 1 (Complex from Homo sapiens)
- Scfv16 (Complex from Mus musculus)
- Cb02-bound cb1r in complex with gi heterotrimer (Complex)
- 3-[(1s)-1-(furan-2-yl)-2-nitro-ethyl]-2-phenyl-1h-indole (332 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
Cryo-EM structure of human receptor with G proteins
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8ikh
Q-score: 0.431
Shen S, Wu C, Lin G, Yang X, Zhou Y, Zhao C, Miao Z, Tian X, Wang K, Yang Z, Liu Z, Guo N, Li Y, Xia A, Zhou P, Liu J, Yan W, Ke B, Yang S, Shao Z
PNAS (2024) 121 pp. e2321532121-e2321532121 [ Pubmed: 38830102 DOI: doi:10.1073/pnas.2321532121 ]
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Scfv16 (28 kDa, Protein from Mus musculus)
- Cannabinoid receptor 1 (35 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Gi heterotrimer (Complex from Homo sapiens)
- Cb05-bound cb1r in complex with gi heterotrimer (Complex)
- 3-[(1r)-1-(2-methoxyphenyl)-2-nitro-ethyl]-2-phenyl-1h-indole (372 Da, Ligand)
- Cannabinoid receptor 1 (Complex from Homo sapiens)
- Scfv16 (Complex from Mus musculus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
Cryo-EM structure of ATP13A2 in the E1-ATP state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8iek
Q-score: 0.377
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Cryo-em structure of atp13a2 in the e1-atp state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the E1-like state
Resolution: 5.65 Å
EM Method: Single-particle
Fitted PDBs: 8iel
Q-score: 0.229
Cryo-EM structure of ATP13A2 in the E2P state
Resolution: 3.35 Å
EM Method: Single-particle
Fitted PDBs: 8iem
Q-score: 0.262
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (Complex from Homo sapiens)
- Polyamine-transporting atpase 13a2 (123 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Spermine (202 Da, Ligand)
- Beryllium trifluoride ion (66 Da, Ligand)
Cryo-EM structure of ATP13A2 in the E2-Pi state
Resolution: 3.25 Å
EM Method: Single-particle
Fitted PDBs: 8ien
Q-score: -0.047
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the e2-pi state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the nominal E1P state
Resolution: 3.78 Å
EM Method: Single-particle
Fitted PDBs: 8ieo
Q-score: 0.312
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the nominal e1p state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the putative of E2 state
Resolution: 4.87 Å
EM Method: Single-particle
Fitted PDBs: 8ier
Q-score: 0.162
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Polyamine-transporting atpase 13a2 (128 kDa, Protein from Homo sapiens)
- Spermine (202 Da, Ligand)
- Cryo-em structure of atp13a2 in the putative of e2 state (Complex from Homo sapiens)
Cryo-EM structure of ATP13A2 in the E1P-ADP state
Resolution: 3.73 Å
EM Method: Single-particle
Fitted PDBs: 8ies
Q-score: 0.308
Mu J, Xue C, Fu L, Yu Z, Nie M, Wu M, Chen X, Liu K, Bu R, Huang Y, Yang B, Han J, Jiang Q, Chan KC, Zhou R, Li H, Huang A, Wang Y, Liu Z
Nat Commun (2023) 14 pp. 1978-1978 [ DOI: doi:10.1038/s41467-023-37741-0 Pubmed: 37031211 ]
- Magnesium ion (24 Da, Ligand)
- Tetrafluoroaluminate ion (102 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Polyamine-transporting atpase 13a2 (125 kDa, Protein from Homo sapiens)
- Cryo-em structure of atp13a2 in the e1p-adp state (Complex from Homo sapiens)
COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex
Resolution: 3.26 Å
EM Method: Single-particle
Fitted PDBs: 7bzf
Q-score: 0.47
Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, Fang X, Yang X, Huang Y, Gao H, Liu F, Ge J, Sun Q, Yang X, Xu W, Liu Z, Yang H, Lou Z, Jiang B, Guddat LW, Gong P, Rao Z
Cell (2020) 182 pp. 417-428.e13 [ DOI: doi:10.1016/j.cell.2020.05.034 Pubmed: 32526208 ]
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (31-mer) (10 kDa, RNA from Foot-and-mouth disease virus)
- Zinc ion (65 Da, Ligand)
- Rna (5'-r(*up*gp*up*up*cp*gp*ap*cp*gp*ap*cp*ap*cp*a)-3') (4 kDa, RNA from Foot-and-mouth disease virus)
- Structure of covid-19 rna-dependent rna polymerase in complex with rna (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)