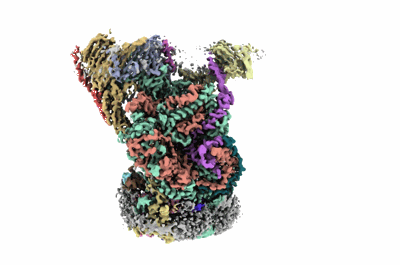
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Resolution: 2.72 Å
EM Method: Single-particle
Fitted PDBs: 8gwk
Q-score: 0.423
Yan L, Huang Y, Ge J, Liu Z, Lu P, Huang B, Gao S, Wang J, Tan L, Ye S, Yu F, Lan W, Xu S, Zhou F, Shi L, Guddat LW, Gao Y, Rao Z, Lou Z
Cell (2022) 185 pp. 4347-4360.e17 [ DOI: doi:10.1016/j.cell.2022.09.037 Pubmed: 36335936 ]
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- E-rtc_rmp-nsp9_gmppnp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Template (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- [(2~{r},3~{s},4~{r},5~{r})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate (371 Da, Ligand)
- Helicase (66 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
- Primer (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8gwm
Q-score: 0.41
Yan L, Huang Y, Ge J, Liu Z, Lu P, Huang B, Gao S, Wang J, Tan L, Ye S, Yu F, Lan W, Xu S, Zhou F, Shi L, Guddat LW, Gao Y, Rao Z, Lou Z
Cell (2022) 185 pp. 4347-4360.e17 [ DOI: doi:10.1016/j.cell.2022.09.037 Pubmed: 36335936 ]
- E-rtc_mmp-nsp9_gmppnp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate) (420 Da, Ligand)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (26-mer) (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (25-mer) (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Helicase (66 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors
Resolution: 3.31 Å
EM Method: Single-particle
Fitted PDBs: 8gw1
Q-score: 0.332
Yan L, Huang Y, Ge J, Liu Z, Lu P, Huang B, Gao S, Wang J, Tan L, Ye S, Yu F, Lan W, Xu S, Zhou F, Shi L, Guddat LW, Gao Y, Rao Z, Lou Z
Cell (2022) 185 pp. 4347-4360.e17 [ DOI: doi:10.1016/j.cell.2022.09.037 Pubmed: 36335936 ]
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- E-rtc_ump-nsp9 (350 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Uridine-5'-monophosphate (324 Da, Ligand)
- Replicase polyprotein 1ab (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Template (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Manganese (ii) ion (54 Da, Ligand)
- Rna (25-mer) (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Helicase (66 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
Cryo-EM structure of the Lhcp complex from Ostreococcus tauri
Resolution: 2.94 Å
EM Method: Single-particle
Fitted PDBs: 8hg3
Q-score: 0.479
Ishii A, Shan J, Sheng X, Kim E, Watanabe A, Yokono M, Noda C, Song C, Murata K, Liu Z, Minagawa J
eLife (2023) 12 [ Pubmed: 36951548 DOI: doi:10.7554/eLife.84488 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Ostreococcus tauri)
- Chlorophyll b (907 Da, Ligand)
- Chlorophyll c2 (608 Da, Ligand)
- Prasinophyte-specific lhc protein (lhcp) complex (Complex from Ostreococcus tauri)
- Chlorophyll a (893 Da, Ligand)
- (1~{s})-3,5,5-trimethyl-4-[(3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol (570 Da, Ligand)
- (3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-1-[(1~{s},4~{s})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one (600 Da, Ligand)
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8hg5
Q-score: 0.486
Ishii A, Shan J, Sheng X, Kim E, Watanabe A, Yokono M, Noda C, Song C, Murata K, Liu Z, Minagawa J
eLife (2023) 12 [ Pubmed: 36951548 DOI: doi:10.7554/eLife.84488 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Ostreococcus tauri)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Ostreococcus tauri)
- Cryo-em structure of the prasinophyte-specific light-harvesting complex (lhcp)from ostreococcus tauri (Complex from Ostreococcus tauri)
- Chlorophyll b (907 Da, Ligand)
- Chlorophyll c2 (608 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
- (1~{s})-3,5,5-trimethyl-4-[(3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol (570 Da, Ligand)
- (3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-1-[(1~{s},4~{s})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one (600 Da, Ligand)
Resolution: 3.44 Å
EM Method: Single-particle
Fitted PDBs: 8hg6
Q-score: 0.412
Ishii A, Shan J, Sheng X, Kim E, Watanabe A, Yokono M, Noda C, Song C, Murata K, Liu Z, Minagawa J
eLife (2023) 12 [ Pubmed: 36951548 DOI: doi:10.7554/eLife.84488 ]
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- Chlorophyll a-b binding protein, chloroplastic (24 kDa, Protein from Ostreococcus tauri)
- Chlorophyll b (907 Da, Ligand)
- Chlorophyll c2 (608 Da, Ligand)
- Prasinophyte-specific lhc protein (lhcp) complex (Complex from Ostreococcus tauri)
- Chlorophyll a (893 Da, Ligand)
- (1~{s})-3,5,5-trimethyl-4-[(3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol (570 Da, Ligand)
- (3~{e},5~{e},7~{e},9~{e},11~{e},13~{e},15~{e},17~{e})-1-[(1~{s},4~{s})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{r},4~{r})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one (600 Da, Ligand)
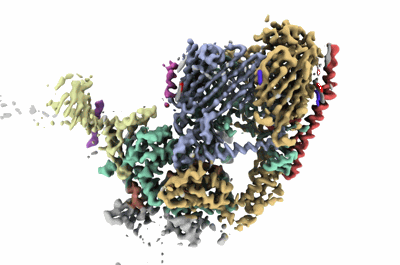
Cryo-EM structure of TOC-TIC supercomplex from Chlamydomonas reinhardtii
Resolution: 2.77 Å
EM Method: Single-particle
Fitted PDBs: 7xzi
Q-score: 0.517
Liu H, Li A, Rochaix JD, Liu Z
Nature (2023) 615 pp. 349-357 [ DOI: doi:10.1038/s41586-023-05744-y Pubmed: 36702157 ]
- Digitonin (1 kDa, Ligand)
- Simp3 (21 kDa, Protein from Chlamydomonas reinhardtii)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- Unknown peptide (1 kDa, Protein from Chlamydomonas reinhardtii)
- Hexadecane (226 Da, Ligand)
- Tic214 (232 kDa, Protein from Chlamydomonas reinhardtii)
- Toc75 (87 kDa, Protein from Chlamydomonas reinhardtii)
- Tic56 (28 kDa, Protein from Chlamydomonas reinhardtii)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Ctap3 (51 kDa, Protein from Chlamydomonas reinhardtii)
- Ctap4 (39 kDa, Protein from Chlamydomonas reinhardtii)
- Ctap5 (42 kDa, Protein from Chlamydomonas reinhardtii)
- Simp2 (13 kDa, Protein from Chlamydomonas reinhardtii)
- Protein tic 20 (29 kDa, Protein from Chlamydomonas reinhardtii)
- Toc34 (44 kDa, Protein from Chlamydomonas reinhardtii)
- Toc90 (102 kDa, Protein from Chlamydomonas reinhardtii)
- Tic100 (105 kDa, Protein from Chlamydomonas reinhardtii)
- Dodecane (170 Da, Ligand)
- A membrane protein complex (812 kDa, Complex from Chlamydomonas reinhardtii)
- Tic15 (14 kDa, Protein from Chlamydomonas reinhardtii)
- Inositol hexakisphosphate (660 Da, Ligand)
Cryo-EM structure of TOC complex from Chlamydomonas reinhardtii.
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 7xzj
Q-score: 0.433
Liu H, Li A, Rochaix JD, Liu Z
Nature (2023) 615 pp. 349-357 [ DOI: doi:10.1038/s41586-023-05744-y Pubmed: 36702157 ]
- Structure of a outer membrane protein complex (Complex from Chlamydomonas reinhardtii)
- Toc75 (87 kDa, Protein from Chlamydomonas reinhardtii)
- Tic214 (232 kDa, Protein from Chlamydomonas reinhardtii)
- Toc34 (44 kDa, Protein from Chlamydomonas reinhardtii)
- Toc90 (102 kDa, Protein from Chlamydomonas reinhardtii)
- Unknown peptide (2 kDa, Protein from Chlamydomonas reinhardtii)
- Toc39 (39 kDa, Protein from Chlamydomonas reinhardtii)
- Tic100 (105 kDa, Protein from Chlamydomonas reinhardtii)
- Inositol hexakisphosphate (660 Da, Ligand)
- Ctap3 (51 kDa, Protein from Chlamydomonas reinhardtii)
A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors
Resolution: 2.66 Å
EM Method: Single-particle
Fitted PDBs: 8gwe
Q-score: 0.019
Yan L, Huang Y, Ge J, Liu Z, Lu P, Huang B, Gao S, Wang J, Tan L, Ye S, Yu F, Lan W, Xu S, Zhou F, Shi L, Guddat LW, Gao Y, Rao Z, Lou Z
Cell (2022) 185 pp. 4347-4360.e17 [ DOI: doi:10.1016/j.cell.2022.09.037 Pubmed: 36335936 ]
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Template (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- E-rtc_rna-nsp9_gmppnp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Phosphoaminophosphonic acid-guanylate ester (522 Da, Ligand)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Rna (5'-r(p*ap*up*up*a)-3') (1 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Helicase nsp13 (65 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Replicase polyprotein 1a (8 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
- Primer (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors
Resolution: 3.39 Å
EM Method: Single-particle
Fitted PDBs: 8gwf
Q-score: 0.29
Yan L, Huang Y, Ge J, Liu Z, Lu P, Huang B, Gao S, Wang J, Tan L, Ye S, Yu F, Lan W, Xu S, Zhou F, Shi L, Guddat LW, Gao Y, Rao Z, Lou Z
Cell (2022) 185 pp. 4347-4360.e17 [ DOI: doi:10.1016/j.cell.2022.09.037 Pubmed: 36335936 ]
- Rna (Complex)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Template (10 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 9 (12 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- E-rtc_rna-nsp9_gtp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Primer (8 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Helicase (66 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
- Nsp (Complex)