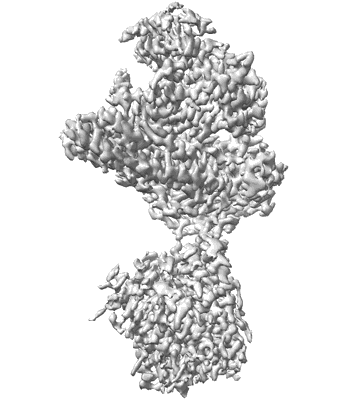
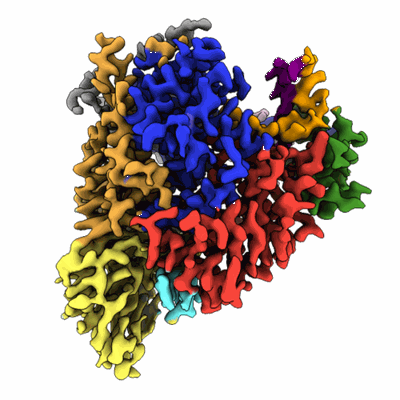
CryoEM structure of human XPR1 in complex with phosphate in state A
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9j51
Q-score: 0.468
Zhang W, Chen Y, Guan Z, Wang Y, Tang M, Du Z, Zhang J, Cheng M, Zuo J, Liu Y, Wang Q, Liu Y, Zhang D, Yin P, Ma L, Liu Z
Nat Commun (2025) 16 pp. 18-18 [ DOI: doi:10.1038/s41467-024-55471-9 Pubmed: 39747008 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Solute carrier family 53 member 1 (84 kDa, Protein from Homo sapiens)
- Human xpr1 (Complex from Homo sapiens)
CryoEM structure of human XPR1 in complex with phosphate in state B
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 9j52
Q-score: 0.468
Zhang W, Chen Y, Guan Z, Wang Y, Tang M, Du Z, Zhang J, Cheng M, Zuo J, Liu Y, Wang Q, Liu Y, Zhang D, Yin P, Ma L, Liu Z
Nat Commun (2025) 16 pp. 18-18 [ DOI: doi:10.1038/s41467-024-55471-9 Pubmed: 39747008 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Solute carrier family 53 member 1 (84 kDa, Protein from Homo sapiens)
- Human xpr1 (Complex from Homo sapiens)
CryoEM structure of human XPR1 in complex with phosphate in state C
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 9j53
Q-score: 0.413
Zhang W, Chen Y, Guan Z, Wang Y, Tang M, Du Z, Zhang J, Cheng M, Zuo J, Liu Y, Wang Q, Liu Y, Zhang D, Yin P, Ma L, Liu Z
Nat Commun (2025) 16 pp. 18-18 [ DOI: doi:10.1038/s41467-024-55471-9 Pubmed: 39747008 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Solute carrier family 53 member 1 (84 kDa, Protein from Homo sapiens)
- Human xpr1 (Complex from Homo sapiens)
CryoEM structure of human XPR1 in apo state
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 9j4x
Q-score: 0.554
Zhang W, Chen Y, Guan Z, Wang Y, Tang M, Du Z, Zhang J, Cheng M, Zuo J, Liu Y, Wang Q, Liu Y, Zhang D, Yin P, Ma L, Liu Z
Nat Commun (2025) 16 pp. 18-18 [ DOI: doi:10.1038/s41467-024-55471-9 Pubmed: 39747008 ]
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Solute carrier family 53 member 1 (84 kDa, Protein from Homo sapiens)
- Human xpr1 (Complex from Homo sapiens)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8z9a
Q-score: 0.54
Wang Y, Qiu L, Wang B, Guan Z, Dong Z, Zhang J, Cao S, Yang L, Wang B, Gong Z, Zhang L, Ma W, Liu Z, Zhang D, Wang G, Yin P
Science (2024) 384 pp. 1453-1460 [ Pubmed: 38870272 DOI: doi:10.1126/science.adn6881 ]
- Odorant receptor, apisorco (52 kDa, Protein from Acyrthosiphon pisum)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Or-orco heterocomplex (Complex from Acyrthosiphon pisum)
- Odorant receptor, apisor5 (42 kDa, Protein from Acyrthosiphon pisum)
- [(2e)-3,7-dimethylocta-2,6-dienyl] ethanoate (196 Da, Ligand)
Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8z9z
Q-score: 0.448
Wang Y, Qiu L, Wang B, Guan Z, Dong Z, Zhang J, Cao S, Yang L, Wang B, Gong Z, Zhang L, Ma W, Liu Z, Zhang D, Wang G, Yin P
Science (2024) 384 pp. 1453-1460 [ Pubmed: 38870272 DOI: doi:10.1126/science.adn6881 ]
- Odorant receptor, apisorco (52 kDa, Protein from Acyrthosiphon pisum)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Odorant receptor, apisor5 (42 kDa, Protein from Acyrthosiphon pisum)
- Or-orco heterocomplex (Complex from Acyrthosiphon pisum)
Cryo-EM structure of VTC complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7ytj
Q-score: 0.485
Guan Z, Chen J, Liu R, Chen Y, Xing Q, Du Z, Cheng M, Hu J, Zhang W, Mei W, Wan B, Wang Q, Zhang J, Cheng P, Cai H, Cao J, Zhang D, Yan J, Yin P, Hothorn M, Liu Z
Nat Commun (2023) 14 pp. 718-718 [ DOI: doi:10.1038/s41467-023-36466-4 Pubmed: 36759618 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Vacuolar transporter chaperone 3 (100 kDa, Protein from Saccharomyces cerevisiae)
- Inositol hexakisphosphate (660 Da, Ligand)
- Vtc complex (Complex from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 4 (86 kDa, Protein from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 1 (15 kDa, Protein from Saccharomyces cerevisiae)
Resolution: 3.85 Å
EM Method: Single-particle
Fitted PDBs: 7v26
Q-score: 0.328
Liu Z, Xu W, Chen Z, Fu W, Zhan W, Gao Y, Zhou J, Zhou Y, Wu J, Wang Q, Zhang X, Hao A, Wu W, Zhang Q, Li Y, Fan K, Chen R, Jiang Q, Mayer CT, Schoofs T, Xie Y, Jiang S, Wen Y, Yuan Z, Wang K, Lu L, Sun L, Wang Q
Protein Cell (2022) 13 pp. 655-675 [ Pubmed: 34554412 DOI: doi:10.1007/s13238-021-00871-6 ]
- Xg005-bound sars-cov-2 s (Complex from Severe acute respiratory syndrome coronavirus 2)
- Xg005 heavy chain (24 kDa, Protein from Homo sapiens)
- Xg005 light chain (22 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 s (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Xg005 (Complex)
SARS-CoV-2 Spike trimer in complex with XG014 Fab
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7v2a
Q-score: 0.482
Liu Z, Xu W, Chen Z, Fu W, Zhan W, Gao Y, Zhou J, Zhou Y, Wu J, Wang Q, Zhang X, Hao A, Wu W, Zhang Q, Li Y, Fan K, Chen R, Jiang Q, Mayer CT, Schoofs T, Xie Y, Jiang S, Wen Y, Yuan Z, Wang K, Lu L, Sun L, Wang Q
Protein Cell (2022) 13 pp. 655-675 [ Pubmed: 34554412 DOI: doi:10.1007/s13238-021-00871-6 ]
- Xg014 in complex with s trimer of sars-cov-2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- The light chain of xg014 fab (22 kDa, Protein from Homo sapiens)
- Xg014 (Complex)
- The heavy chain of xg014 (25 kDa, Protein from Homo sapiens)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- S trimer of sars-cov-2 (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex
Resolution: 3.26 Å
EM Method: Single-particle
Fitted PDBs: 7bzf
Q-score: 0.47
Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, Fang X, Yang X, Huang Y, Gao H, Liu F, Ge J, Sun Q, Yang X, Xu W, Liu Z, Yang H, Lou Z, Jiang B, Guddat LW, Gong P, Rao Z
Cell (2020) 182 pp. 417-428.e13 [ DOI: doi:10.1016/j.cell.2020.05.034 Pubmed: 32526208 ]
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (31-mer) (10 kDa, RNA from Foot-and-mouth disease virus)
- Zinc ion (65 Da, Ligand)
- Rna (5'-r(*up*gp*up*up*cp*gp*ap*cp*gp*ap*cp*ap*cp*a)-3') (4 kDa, RNA from Foot-and-mouth disease virus)
- Structure of covid-19 rna-dependent rna polymerase in complex with rna (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)