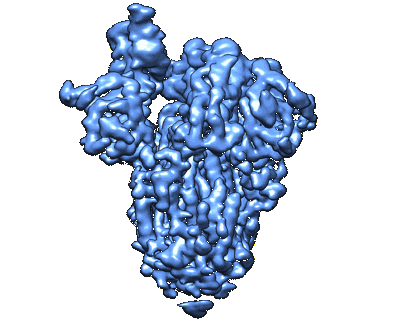
Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer)
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 7kvc
Q-score: 0.305
Llauger G, Melero R, Monti D, Sycz G, Huck-Iriart C, Cerutti ML, Klinke S, Mikkelsen E, Tijman A, Arranz R, Alfonso V, Arellano SM, Goldbaum FA, Sterckx YGJ, Carazo JM, Kaufman SB, Dans PD, Del Vas M, Otero LH
Mbio (2023) 14 pp. e0002323-e0002323 [ Pubmed: 36786587 DOI: doi:10.1128/mbio.00023-23 ]
Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer)
Resolution: 6.8 Å
EM Method: Single-particle
Fitted PDBs: 7kvd
Q-score: 0.211
Llauger G, Melero R, Monti D, Sycz G, Huck-Iriart C, Cerutti ML, Klinke S, Mikkelsen E, Tijman A, Arranz R, Alfonso V, Arellano SM, Goldbaum FA, Sterckx YGJ, Carazo JM, Kaufman SB, Dans PD, Del Vas M, Otero LH
Mbio (2023) 14 pp. e0002323-e0002323 [ Pubmed: 36786587 DOI: doi:10.1128/mbio.00023-23 ]
SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7qdg
Q-score: 0.31
Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Ruiz-Rodriguez P, Lopez-Redondo ML, Frances-Gomez C, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Arranz R, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM
PLoS Pathog (2022) 18 pp. e1010631-e1010631 [ DOI: doi:10.1371/journal.ppat.1010631 Pubmed: 35816514 ]
SARS-CoV-2 S protein S:D614G mutant 1-up
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 7qdh
Q-score: 0.079
Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Ruiz-Rodriguez P, Lopez-Redondo ML, Frances-Gomez C, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Arranz R, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM
PLoS Pathog (2022) 18 pp. e1010631-e1010631 [ DOI: doi:10.1371/journal.ppat.1010631 Pubmed: 35816514 ]
- Sars-cov-2 s protein s:d614g mutant (340 kDa, Complex from Severe acute respiratory syndrome-related coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein,fibritin (138 kDa, Protein from Escherichia virus T4)
SARS-CoV-2 spike in prefusion state
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 6zow
Q-score: 0.451
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Water (18 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Spike homotrimer in prefusion state (Complex from Severe acute respiratory syndrome coronavirus 2)
- Dimethyl sulfoxide (78 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 6zp5
Q-score: 0.417
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Dimethyl sulfoxide (78 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation)
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6zp7
Q-score: 0.368
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Sars-cov-2 spike in prefusion state (flexibility analysis, 1-up open conformation) (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Dimethyl sulfoxide (78 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)