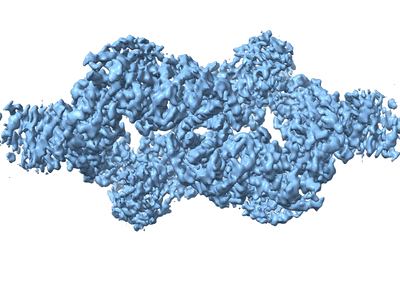
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8p99
Q-score: 0.343
Gargantilla M, Frances C, Adhav A, Forcada-Nadal A, Martinez-Gualda B, Marti-Mari O, Lopez-Redondo ML, Melero R, Marco-Marin C, Gougeard N, Espinosa C, Rubio-Del-Campo A, Ruiz-Partida R, Hernandez-Sierra MDP, Villamayor-Belinchon L, Bravo J, Llacer JL, Marina A, Rubio V, San-Felix A, Geller R, Perez-Perez MJ
J Med Chem (2023) 66 pp. 10432-10457 [ DOI: doi:10.1021/acs.jmedchem.3c00576 Pubmed: 37471688 ]
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 8p9y
Q-score: 0.117
Gargantilla M, Frances C, Adhav A, Forcada-Nadal A, Martinez-Gualda B, Marti-Mari O, Lopez-Redondo ML, Melero R, Marco-Marin C, Gougeard N, Espinosa C, Rubio-Del-Campo A, Ruiz-Partida R, Hernandez-Sierra MDP, Villamayor-Belinchon L, Bravo J, Llacer JL, Marina A, Rubio V, San-Felix A, Geller R, Perez-Perez MJ
J Med Chem (2023) 66 pp. 10432-10457 [ DOI: doi:10.1021/acs.jmedchem.3c00576 Pubmed: 37471688 ]
- Spike glycoprotein - severe acute respiratory syndrome coronavirus 2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s1,spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sodium ion (22 Da, Ligand)
- [(2~{s})-2-[[4-(2-azanylethanoylamino)-7-[[(2~{s})-3-[2-(4-nitrophenyl)sulfanyl-1~{h}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-4-[3-[[(2~{s})-3-[2-(4-nitrophenyl)sulfanyl-1~{h}-indol-3-yl]-1-oxidanylidene-1-sodiooxy-propan-2-yl]amino]-3-oxidanylidene-propyl]-7-oxidanylidene-heptanoyl]amino]-3-[2-(4-nitrophenyl)sulfanyl-1~{h}-indol-3-yl]propanoyl]oxysodium (1 kDa, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7r4i
Q-score: 0.241
Casasnovas JM, Margolles Y, Noriega MA, Guzman M, Arranz R, Melero R, Casanova M, Corbera JA, Jimenez-de-Oya N, Gastaminza P, Garaigorta U, Saiz JC, Martin-Acebes MA, Fernandez LA
Front Immunol (2022) 13 pp. 863831-863831 [ Pubmed: 35547740 DOI: doi:10.3389/fimmu.2022.863831 ]
- Camel-derived nanobody 2.15 (13 kDa, Protein from Camelus dromedarius)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- Nanobody 2.15 (Complex from Camelus dromedarius)
- The trimeric sars-coov-2 spike in complex with the 2.15 neutralizing nanobody (10 MDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7r4q
Q-score: -0.014
Casasnovas JM, Margolles Y, Noriega MA, Guzman M, Arranz R, Melero R, Casanova M, Corbera JA, Jimenez-de-Oya N, Gastaminza P, Garaigorta U, Saiz JC, Martin-Acebes MA, Fernandez LA
Front Immunol (2022) 13 pp. 863831-863831 [ Pubmed: 35547740 DOI: doi:10.3389/fimmu.2022.863831 ]
- Nanobody 1.29 (Complex from Camelus dromedarius)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Camel-derived nanobody 1.29 (13 kDa, Protein from Camelus dromedarius)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- The trimeric sars-coov-2 spike in complex with the 1.29 neutralizing nanobody (10 MDa, Complex from Severe acute respiratory syndrome coronavirus 2)
The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 7r4r
Q-score: 0.03
Casasnovas JM, Margolles Y, Noriega MA, Guzman M, Arranz R, Melero R, Casanova M, Corbera JA, Jimenez-de-Oya N, Gastaminza P, Garaigorta U, Saiz JC, Martin-Acebes MA, Fernandez LA
Front Immunol (2022) 13 pp. 863831-863831 [ Pubmed: 35547740 DOI: doi:10.3389/fimmu.2022.863831 ]
- The trimeric sars-coov-2 spike in complex with the 1.10 neutralizing nanobody (10 MDa, Complex)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Camel-derived nanobody 1.10 (13 kDa, Protein from Camelus dromedarius)
- Sars-cov-2 spike (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Nanobody 1.10 (Complex from Camelus dromedarius)
Resolution: 4.19 Å
EM Method: Single-particle
Fitted PDBs: 7a1d
Q-score: 0.281
Lazaro M, Melero R, Huet C, Lopez-Alonso JP, Delgado S, Dodu A, Bruch EM, Abriata LA, Alzari PM, Valle M, Lisa MN
Commun Biol (2021) 4 pp. 684-684 [ Pubmed: 34083757 DOI: doi:10.1038/s42003-021-02222-x ]
- Cryo-em map of the large glutamate dehydrogenase composed of 180 kda subunits from mycobacterium smegmatis (open conformation) (705 kDa, Complex from Mycolicibacterium smegmatis MC2 155)
- Nad-specific glutamate dehydrogenase (176 kDa, Protein from Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155))
SARS-CoV-2 spike in prefusion state
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 6zow
Q-score: 0.451
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Water (18 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Spike homotrimer in prefusion state (Complex from Severe acute respiratory syndrome coronavirus 2)
- Dimethyl sulfoxide (78 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation)
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 6zp5
Q-score: 0.417
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Dimethyl sulfoxide (78 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation)
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 6zp7
Q-score: 0.368
Melero R, Sorzano COS, Foster B, Vilas JL, Martinez M, Marabini R, Ramirez-Aportela E, Sanchez-Garcia R, Herreros D, Del Cano L, Losana P, Fonseca-Reyna YC, Conesa P, Wrapp D, Chacon P, McLellan JS, Tagare HD, Carazo JM
IUCrJ (2020) 7 pp. 1059-1069 [ DOI: doi:10.1107/S2052252520012725 Pubmed: 33063791 ]
- Sars-cov-2 spike in prefusion state (flexibility analysis, 1-up open conformation) (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Alpha-d-mannopyranose (180 Da, Ligand)
- Dimethyl sulfoxide (78 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)