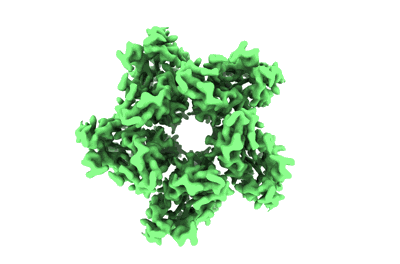
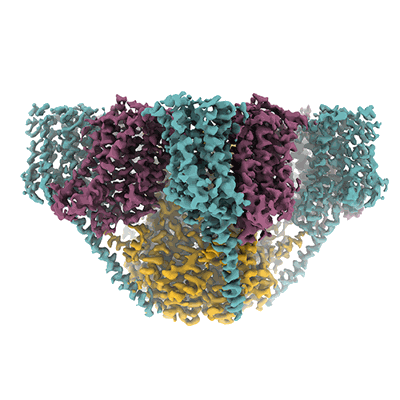
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Gi heterotrimer
Resolution: 4.9 Å
EM Method: Single-particle
Fitted PDBs: 9epr
Q-score: 0.248
Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ
Nat Commun (2024) 15 pp. 8928-8928 [ Pubmed: 39414813 DOI: doi:10.1038/s41467-024-53208-2 ]
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(t) subunit gamma-t1 (8 kDa, Protein from Bos taurus)
- Retinal (284 Da, Ligand)
- Kumopsin1 (42 kDa, Protein from Hasarius adansoni)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (Complex from Homo sapiens)
- Ternary complex of jumping spider rhodopsin-1 with a human gi heterotrimer (Complex)
- Jumping spider rhodopsin-1 (Complex from Hasarius adansoni)
- Guanine nucleotide-binding protein g(t) subunit gamma-t1 (Complex from Bos taurus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (Complex from Bos taurus)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Bos taurus)
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Resolution: 4.06 Å
EM Method: Single-particle
Fitted PDBs: 9epp
Q-score: 0.413
Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ
Nat Commun (2024) 15 pp. 8928-8928 [ Pubmed: 39414813 DOI: doi:10.1038/s41467-024-53208-2 ]
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (6 kDa, Protein from Homo sapiens)
- 11,20-ethanoretinal (310 Da, Ligand)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Ternary complex of jumping spider rhodopsin-1 with a chimeric giq heterotrimer (Complex)
- Jumping spider rhodopsin-1 (Complex from Hasarius adansoni)
- Kumopsin1 (37 kDa, Protein from Hasarius adansoni)
- Human gi/jumping spider gq chimera (Complex from Hasarius adansoni)
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Resolution: 4.15 Å
EM Method: Single-particle
Fitted PDBs: 9epq
Q-score: 0.348
Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ
Nat Commun (2024) 15 pp. 8928-8928 [ Pubmed: 39414813 DOI: doi:10.1038/s41467-024-53208-2 ]
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (6 kDa, Protein from Homo sapiens)
- 11,20-ethanoretinal (310 Da, Ligand)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Ternary complex of jumping spider rhodopsin-1 with a chimeric giq heterotrimer (Complex)
- Jumping spider rhodopsin-1 (Complex from Hasarius adansoni)
- Kumopsin1 (37 kDa, Protein from Hasarius adansoni)
- Human gi/jumping spider gq chimera (Complex from Hasarius adansoni)
The potassium-selective channelrhodopsin HcKCR1 and HcKCR2 in lipid nanodisc
Tajima S, Kim YS, Fukuda M, Jo Y, Wang PY, Paggi JM, Inoue M, Byrne EFX, Kishi KE, Nakamura S, Ramakrishnan C, Takaramoto S, Nagata T, Konno M, Sugiura M, Katayama K, Matsui TE, Yamashita K, Kim S, Ikeda H, Kim J, Kandori H, Dror RO, Inoue K, Deisseroth K, Kato HE
Cell (2023) [ Pubmed: 37652010 DOI: 10.1016/j.cell.2023.08.009 ]
Image sets:
Resolution: 2.56 Å
EM Method: Single-particle
Fitted PDBs: 8h86
Q-score: 0.661
Tajima S, Kim YS, Fukuda M, Jo Y, Wang PY, Paggi JM, Inoue M, Byrne EFX, Kishi KE, Nakamura S, Ramakrishnan C, Takaramoto S, Nagata T, Konno M, Sugiura M, Katayama K, Matsui TE, Yamashita K, Kim S, Ikeda H, Kim J, Kandori H, Dror RO, Inoue K, Deisseroth K, Kato HE
Cell (2023) 186 pp. 4325-4344.e26 [ DOI: doi:10.1016/j.cell.2023.08.009 Pubmed: 37652010 ]
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Retinal (284 Da, Ligand)
- Hckcr1 (Complex from Hyphochytrium catenoides)
- (7r,17e,20e)-4-hydroxy-n,n,n-trimethyl-9-oxo-7-[(palmitoyloxy)methyl]-3,5,8-trioxa-4-phosphahexacosa-17,20-dien-1-aminium 4-oxide (759 Da, Ligand)
- Hckcr1 (31 kDa, Protein from Hyphochytrium catenoides)
Resolution: 2.53 Å
EM Method: Single-particle
Fitted PDBs: 8h87
Q-score: 0.682
Tajima S, Kim YS, Fukuda M, Jo Y, Wang PY, Paggi JM, Inoue M, Byrne EFX, Kishi KE, Nakamura S, Ramakrishnan C, Takaramoto S, Nagata T, Konno M, Sugiura M, Katayama K, Matsui TE, Yamashita K, Kim S, Ikeda H, Kim J, Kandori H, Dror RO, Inoue K, Deisseroth K, Kato HE
Cell (2023) 186 pp. 4325-4344.e26 [ DOI: doi:10.1016/j.cell.2023.08.009 Pubmed: 37652010 ]
- Water (18 Da, Ligand)
- Retinal (284 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- (7r,17e,20e)-4-hydroxy-n,n,n-trimethyl-9-oxo-7-[(palmitoyloxy)methyl]-3,5,8-trioxa-4-phosphahexacosa-17,20-dien-1-aminium 4-oxide (759 Da, Ligand)
- Hckcr2 (Complex from Hyphochytrium catenoides)
- Hckcr2 (34 kDa, Protein from Hyphochytrium catenoides)
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc
Resolution: 2.66 Å
EM Method: Single-particle
Fitted PDBs: 8iu0
Q-score: 0.636
Tajima S, Kim YS, Fukuda M, Jo Y, Wang PY, Paggi JM, Inoue M, Byrne EFX, Kishi KE, Nakamura S, Ramakrishnan C, Takaramoto S, Nagata T, Konno M, Sugiura M, Katayama K, Matsui TE, Yamashita K, Kim S, Ikeda H, Kim J, Kandori H, Dror RO, Inoue K, Deisseroth K, Kato HE
Cell (2023) 186 pp. 4325-4344.e26 [ DOI: doi:10.1016/j.cell.2023.08.009 Pubmed: 37652010 ]
- Water (18 Da, Ligand)
- Palmitic acid (256 Da, Ligand)
- Retinal (284 Da, Ligand)
- Hckcr1 (Complex from Hyphochytrium catenoides)
- (7r,17e,20e)-4-hydroxy-n,n,n-trimethyl-9-oxo-7-[(palmitoyloxy)methyl]-3,5,8-trioxa-4-phosphahexacosa-17,20-dien-1-aminium 4-oxide (759 Da, Ligand)
- Hckcr1 (31 kDa, Protein from Hyphochytrium catenoides)
Cryo-EM structure of the zeaxanthin-bound kin4B8
Resolution: 2.3 Å
EM Method: Single-particle
Fitted PDBs: 8i2z
Q-score: 0.703
Chazan A, Das I, Fujiwara T, Murakoshi S, Rozenberg A, Molina-Marquez A, Sano FK, Tanaka T, Gomez-Villegas P, Larom S, Pushkarev A, Malakar P, Hasegawa M, Tsukamoto Y, Ishizuka T, Konno M, Nagata T, Mizuno Y, Katayama K, Abe-Yoshizumi R, Ruhman S, Inoue K, Kandori H, Leon R, Shihoya W, Yoshizawa S, Sheves M, Nureki O, Beja O
Nature (2023) 615 pp. 535-540 [ DOI: doi:10.1038/s41586-023-05774-6 Pubmed: 36859551 ]
- Xanthorhodopsin (28 kDa, Protein from uncultured Bdellovibrionales bacterium)
- Water (18 Da, Ligand)
- Retinal (284 Da, Ligand)
- Kin4b8 (Cellular component from uncultured Bdellovibrionales bacterium)
- Zeaxanthin (568 Da, Ligand)
Cryo-EM structure of Bestrhodopsin (rhodopsin-rhodopsin-bestrophin) complex
Resolution: 3.21 Å
EM Method: Single-particle
Fitted PDBs: 7pl9
Q-score: 0.458
Rozenberg A, Kaczmarczyk I, Matzov D, Vierock J, Nagata T, Sugiura M, Katayama K, Kawasaki Y, Konno M, Nagasaka Y, Aoyama M, Das I, Pahima E, Church J, Adam S, Borin VA, Chazan A, Augustin S, Wietek J, Dine J, Peleg Y, Kawanabe A, Fujiwara Y, Yizhar O, Sheves M, Schapiro I, Furutani Y, Kandori H, Inoue K, Hegemann P, Beja O, Shalev-Benami M
Nat Struct Mol Biol (2022) 29 pp. 592-603 [ Pubmed: 35710843 DOI: doi:10.1038/s41594-022-00783-x ]
- Rhodopsin (670 kDa, Complex from Phaeocystis antarctica)
- Rhodopsin (134 kDa, Protein from Phaeocystis)
- Retinal (284 Da, Ligand)
The pump-like chanelrhodopsin ChRmine
Kishi KE, Kim YS, Fukuda M, Inoue M, Kusakizako T, Wang PY, Ramakrishnan C, Byrne EFX, Thadhani E, Paggi JM, Matsui TE, Yamashita K, Nagata T, Konno M, Quirin S, Lo M, Benster T, Uemura T, Liu K, Shibata M, Nomura N, Iwata S, Nureki O, Dror RO, Inoue K, Deisseroth K, Kato HE
Cell (2022) [ DOI: 10.1016/j.cell.2022.01.007 Pubmed: 35114111 ]
Image sets: