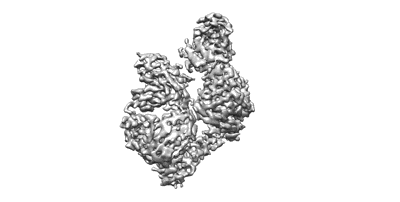
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)
Resolution: 3.23 Å
EM Method: Single-particle
Fitted PDBs: 8yf6
Q-score: 0.394
Sterbova P, Wang CH, Carillo KJD, Lou YC, Kato T, Namba K, Tzou DM, Chang WH
Acs Infect Dis. (2024) 10 pp. 3304-3319 [ DOI: doi:10.1021/acsinfecdis.4c00407 Pubmed: 39087906 ]
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)
Resolution: 2.82 Å
EM Method: Single-particle
Fitted PDBs: 8yf7
Q-score: 0.364
Sterbova P, Wang CH, Carillo KJD, Lou YC, Kato T, Namba K, Tzou DM, Chang WH
Acs Infect Dis. (2024) 10 pp. 3304-3319 [ DOI: doi:10.1021/acsinfecdis.4c00407 Pubmed: 39087906 ]
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 8yf8
Q-score: 0.321
Sterbova P, Wang CH, Carillo KJD, Lou YC, Kato T, Namba K, Tzou DM, Chang WH
Acs Infect Dis. (2024) 10 pp. 3304-3319 [ DOI: doi:10.1021/acsinfecdis.4c00407 Pubmed: 39087906 ]
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 8yf9
Q-score: 0.317
Sterbova P, Wang CH, Carillo KJD, Lou YC, Kato T, Namba K, Tzou DM, Chang WH
Acs Infect Dis. (2024) 10 pp. 3304-3319 [ DOI: doi:10.1021/acsinfecdis.4c00407 Pubmed: 39087906 ]
Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2
Resolution: 3.63 Å
EM Method: Single-particle
Fitted PDBs: 8hlb
Q-score: 0.446
Akiba H, Fujita J, Ise T, Nishiyama K, Miyata T, Kato T, Namba K, Ohno H, Kamada H, Nagata S, Tsumoto K
Commun Biol (2023) 6 pp. 987-987 [ DOI: doi:10.1038/s42003-023-05326-8 Pubmed: 37758868 ]
- Tr92 light chain (23 kDa, Protein from Homo sapiens)
- Cryo-em structure of biparatopic antibody bp109-92 in complex with tnfr2 (Complex from Homo sapiens)
- Tr109 light chain (23 kDa, Protein from Homo sapiens)
- Tumor necrosis factor receptor superfamily member 1b,maltose/maltodextrin-binding periplasmic protein (60 kDa, Protein from Escherichia coli (strain K12))
- Tr109 heavy chain (25 kDa, Protein from Homo sapiens)
- Tr92 heavy chain (25 kDa, Protein from Homo sapiens)
Cryo-EM structure of KpFtsZ-monobody double helical tube
Resolution: 2.67 Å
EM Method: Helical reconstruction
Fitted PDBs: 8h1o
Q-score: 0.621
Fujita J, Amesaka H, Yoshizawa T, Hibino K, Kamimura N, Kuroda N, Konishi T, Kato Y, Hara M, Inoue T, Namba K, Tanaka SI, Matsumura H
Nat Commun (2023) 14 pp. 4073-4073 [ Pubmed: 37429870 DOI: doi:10.1038/s41467-023-39807-5 ]
- Guanosine-5'-diphosphate (443 Da, Ligand)
- Monobody (Complex from Homo sapiens)
- Kpftsz (Complex from Klebsiella pneumoniae)
- Cryo-em structure of kpftsz-monobody double helical tube (Complex)
- Mb(ec/kpftsz_s1) (9 kDa, Protein from Homo sapiens)
- Cell division protein ftsz (40 kDa, Protein from Klebsiella pneumoniae)
Cryo-EM structure of KpFtsZ single filament
Resolution: 3.03 Å
EM Method: Helical reconstruction
Fitted PDBs: 8ibn
Q-score: 0.36
Fujita J, Amesaka H, Yoshizawa T, Hibino K, Kamimura N, Kuroda N, Konishi T, Kato Y, Hara M, Inoue T, Namba K, Tanaka SI, Matsumura H
Nat Commun (2023) 14 pp. 4073-4073 [ Pubmed: 37429870 DOI: doi:10.1038/s41467-023-39807-5 ]
- Cryo-em structure of kpftsz single filament (Complex)
- Kpftsz (Complex from Klebsiella pneumoniae)
- Potassium ion (39 Da, Ligand)
- Phosphomethylphosphonic acid guanylate ester (521 Da, Ligand)
- Cell division protein ftsz (40 kDa, Protein from Klebsiella pneumoniae)
34-fold symmetry Salmonella S ring formed by full-length FliF
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7d84
Q-score: 0.447
Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K
Nat Commun (2021) 12 pp. 4223-4223 [ DOI: doi:10.1038/s41467-021-24507-9 Pubmed: 34244518 ]
Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 6lu1
Q-score: 0.582
Coruh O, Frank A, Tanaka H, Kawamoto A, El-Mohsnawy E, Kato T, Namba K, Gerle C, Nowaczyk MM, Kurisu G
Commun Biol (2021) 4 pp. 304-304 [ Pubmed: 33686186 DOI: doi:10.1038/s42003-021-01808-9 ]
- Photosystem i p700 chlorophyll a apoprotein a1 (83 kDa, Protein from Thermosynechococcus elongatus BP-1)
- Photosystem i reaction center subunit ii (15 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Photosystem i p700 chlorophyll a apoprotein a2 (83 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Photosystem i reaction center subunit ix (4 kDa, Protein from Thermosynechococcus elongatus BP-1)
- Cyanobacterial psi monomer from t. elongatus (350 MDa, Complex from Thermosynechococcus elongatus BP-1)
- Photosystem i reaction center subunit xii (3 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Water (18 Da, Ligand)
- Photosystem i reaction center subunit xi (16 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Photosystem i reaction center subunit iv (8 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Photosystem i reaction center subunit viii (4 kDa, Protein from Thermosynechococcus elongatus BP-1)
- Beta-carotene (536 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Phylloquinone (450 Da, Ligand)
- 1,2-distearoyl-monogalactosyl-diglyceride (787 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Photosystem i reaction center subunit iii (17 kDa, Protein from Thermosynechococcus elongatus BP-1)
- Photosystem i iron-sulfur center (8 kDa, Protein from Thermosynechococcus elongatus (strain BP-1))
- Chlorophyll a (893 Da, Ligand)