SARS-CoV-2 Spike S2 bound to Fab 54043-5
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8vcr
Q-score: 0.535
Johnson NV, Wall SC, Kramer KJ, Holt CM, Periasamy S, Richardson SI, Manamela NP, Suryadevara N, Andreano E, Paciello I, Pierleoni G, Piccini G, Huang Y, Ge P, Allen JD, Uno N, Shiakolas AR, Pilewski KA, Nargi RS, Sutton RE, Abu-Shmais AA, Parks R, Haynes BF, Carnahan RH, Crowe Jr JE, Montomoli E, Rappuoli R, Bukreyev A, Ross TM, Sautto GA, McLellan JS, Georgiev IS
Structure (2024) 32 pp. 1893- [ Pubmed: 39326419 DOI: doi:10.1016/j.str.2024.08.022 ]
- S2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s2 (65 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab 54043-5 (Complex from Homo sapiens)
- 54043-5 fab light chain (23 kDa, Protein from Homo sapiens)
- Trimeric s2 bound to fab 54043-5 (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 54043-5 fab heavy chain (25 kDa, Protein from Homo sapiens)
SARS-CoV-2 S-RBD + Fab 54042-4
Resolution: 2.69 Å
EM Method: Single-particle
Fitted PDBs: 7t01
Q-score: 0.463
Kramer KJ, Johnson NV, Shiakolas AR, Suryadevara N, Periasamy S, Raju N, Williams JK, Wrapp D, Zost SJ, Walker LM, Wall SC, Holt CM, Hsieh CL, Sutton RE, Paulo A, Nargi RS, Davidson E, Doranz BJ, Crowe Jr JE, Bukreyev A, Carnahan RH, McLellan JS, Georgiev IS
Cell Rep (2021) 37 pp. 109784-109784 [ DOI: doi:10.1016/j.celrep.2021.109784 Pubmed: 34592170 ]
- 54042-4 fab - heavy chain (13 kDa, Protein from Homo sapiens)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 54042-4 fab - light chain (11 kDa, Protein from Homo sapiens)
- Complex of sars-cov-2 s-ecd with 54042-4 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-143)
Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe JE
Cell (2020) 183 pp. 1884-1.9e+26 [ DOI: 10.1016/j.cell.2020.11.011 Pubmed: 33301709 ]
Image sets:
Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-33)
Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe JE
Cell (2020) 183 pp. 1884-1900.e23 [ DOI: 10.1016/j.cell.2020.11.011 Pubmed: 33301709 ]
Image sets:
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2489 (NTD)
Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, Nargi RS, Sutton RE, Winkler ES, Chen EC, Fouch ME, Davidson E, Doranz BJ, Carnahan RH, Thackray LB, Diamond MS, Crowe JE
bioRxiv (2021) [ Pubmed: 33501445 DOI: doi:10.1101/2021.01.19.427324 ]
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2676 (NTD)
Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, Nargi RS, Sutton RE, Winkler ES, Chen EC, Fouch ME, Davidson E, Doranz BJ, Carnahan RH, Thackray LB, Diamond MS, Crowe JE
bioRxiv (2021) [ Pubmed: 33501445 DOI: doi:10.1101/2021.01.19.427324 ]
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-33 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
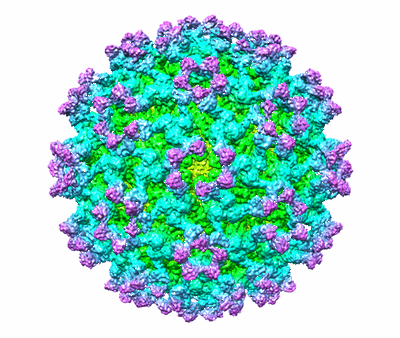
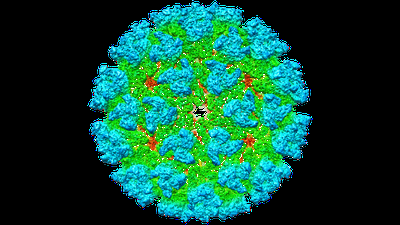
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6xo4
Q-score: 0.343
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
- Eastern equine encephalitis virus (Virus from Eastern equine encephalitis virus)
- Togavirin (29 kDa, Protein from Eastern equine encephalitis virus)
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
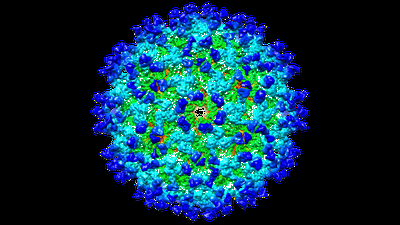
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP with Fab EEEV-143.
Resolution: 8.5 Å
EM Method: Single-particle
Fitted PDBs: 6xob
Q-score: 0.182
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Eeev-143 fab heavy chain (23 kDa, Protein from Homo sapiens)
- Togavirin (29 kDa, Protein from Eastern equine encephalitis virus)
- Togavirin (47 kDa, Protein from Eastern equine encephalitis virus)
- Eastern equine encephalitis virus (Virus from Eastern equine encephalitis virus)
- Eeev-143 fab light chain (23 kDa, Protein from Homo sapiens)