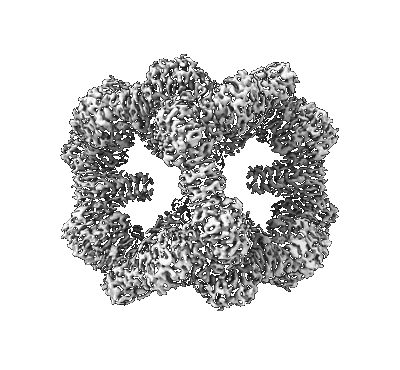
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8szz
Q-score: 0.591
Li Z, Wang S, Nattermann U, Bera AK, Borst AJ, Yaman MY, Bick MJ, Yang EC, Sheffler W, Lee B, Seifert S, Hura GL, Nguyen H, Kang A, Dalal R, Lubner JM, Hsia Y, Haddox H, Courbet A, Dowling Q, Miranda M, Favor A, Etemadi A, Edman NI, Yang W, Weidle C, Sankaran B, Negahdari B, Ross MB, Ginger DS, Baker D
Nat Mater (2023) 22 pp. 1556-1563 [ Pubmed: 37845322 DOI: doi:10.1038/s41563-023-01683-1 ]
- O32-zl4 component a (8 kDa, Protein from synthetic construct)
- O32-zl4 (764 kDa, Complex from Thermotoga maritima)
- O32-zl4 component b (23 kDa, Protein from Thermotoga maritima)
- Sodium ion (22 Da, Ligand)
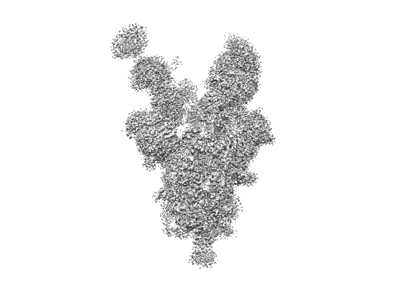
SARS-CoV-2 spike trimer in complex with Fab NE12, ensemble map
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Sars-cov-2 s6p spike trimer (Protein from Severe acute respiratory syndrome coronavirus 2)
- Ne12 fab heavy chain (Protein from Homo sapiens)
- Ne12 fab light chain (Protein from Homo sapiens)
- Sars-cov-2 s6p spike trimer in complex with fab ne12 (580 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
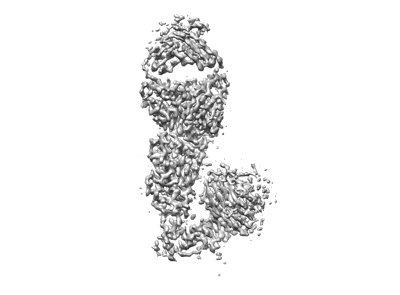
SARS-CoV-2 spike trimer in complex with Fab NE12, local refinement map
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7u9o
Q-score: 0.518
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Ne12 fab light chain (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike receptor-binding domain in complex with fab ne12 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Ne12 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
SARS-CoV-2 spike trimer in complex with Fab NA8, ensemble map
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Sars-cov-2 s6p spike trimer in complex with fab na8 (580 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Na8 fab light chain (Protein from Homo sapiens)
- Sars-cov-2 s6p spike trimer (Protein from Severe acute respiratory syndrome coronavirus 2)
- Na8 fab heavy chain (Protein from Homo sapiens)
SARS-CoV-2 spike trimer in complex with Fab NA8, local refinement map
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7u9p
Q-score: 0.528
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Sars-cov-2 spike receptor-binding domain in complex with fab na8 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Na8 fab light chain (23 kDa, Protein from Homo sapiens)
- Na8 fab heavy chain (24 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
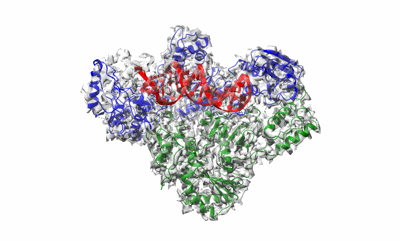
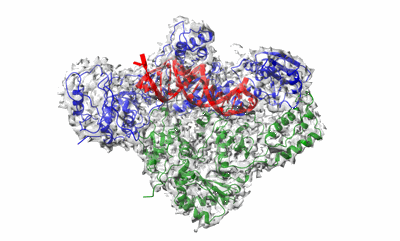
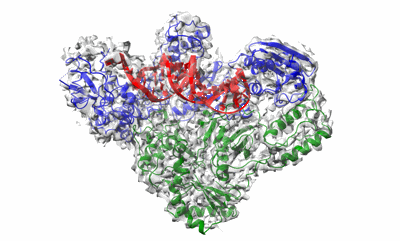
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine
Resolution: 3.32 Å
EM Method: Single-particle
Fitted PDBs: 7z24
Q-score: 0.494
Singh AK, De Wijngaert B, Bijnens M, Uyttersprot K, Nguyen H, Martinez SE, Schols D, Herdewijn P, Pannecouque C, Arnold E, Das K
PNAS (2022) 119 pp. e2203660119-e2203660119 [ Pubmed: 35858448 DOI: doi:10.1073/pnas.2203660119 ]
- Hiv-1 reverse transcriptase p51 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- Hiv-1 reverse transcriptase with a dna aptamer in complex with nevirapine (Complex from Human immunodeficiency virus type 1 BH10)
- Dna (38-mer) (11 kDa, DNA from synthetic construct)
- Hiv-1 reverse transcriptase p66 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (64 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (50 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- 11-cyclopropyl-5,11-dihydro-4-methyl-6h-dipyrido[3,2-b:2',3'-e][1,4]diazepin-6-one (266 Da, Ligand)
Resolution: 3.38 Å
EM Method: Single-particle
Fitted PDBs: 7z29
Q-score: 0.463
Singh AK, De Wijngaert B, Bijnens M, Uyttersprot K, Nguyen H, Martinez SE, Schols D, Herdewijn P, Pannecouque C, Arnold E, Das K
PNAS (2022) 119 pp. e2203660119-e2203660119 [ Pubmed: 35858448 DOI: doi:10.1073/pnas.2203660119 ]
- Dna (38-mer) (11 kDa, DNA from synthetic construct)
- Hiv-1 reverse transcriptase p51 subunit with e138k mutation (Complex from Human immunodeficiency virus type 1 BH10)
- Nnrti resistant m184i/e138k mutant hiv-1 reverse transcriptase with a dna aptamer in complex with nevirapine (Complex from Human immunodeficiency virus type 1 BH10)
- Hiv-1 reverse transcriptase p66 subunit with m184i mutation (Complex from Human immunodeficiency virus type 1 BH10)
- P51 rt (50 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (64 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- 11-cyclopropyl-5,11-dihydro-4-methyl-6h-dipyrido[3,2-b:2',3'-e][1,4]diazepin-6-one (266 Da, Ligand)
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with rilpivirine
Resolution: 3.38 Å
EM Method: Single-particle
Fitted PDBs: 7z2d
Q-score: 0.452
Singh AK, De Wijngaert B, Bijnens M, Uyttersprot K, Nguyen H, Martinez SE, Schols D, Herdewijn P, Pannecouque C, Arnold E, Das K
PNAS (2022) 119 pp. e2203660119-e2203660119 [ Pubmed: 35858448 DOI: doi:10.1073/pnas.2203660119 ]
- Hiv-1 reverse transcriptase p51 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- 4-{[4-({4-[(e)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile (366 Da, Ligand)
- Hiv-1 reverse transcriptase p66 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (64 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (50 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Dna (38-mer) (11 kDa, DNA from synthetic construct)
- Hiv-1 reverse transcriptase with a dna aptamer in complex with rilpivirine (Complex from Human immunodeficiency virus type 1 BH10)
Resolution: 3.45 Å
EM Method: Single-particle
Fitted PDBs: 7z2e
Q-score: 0.448
Singh AK, De Wijngaert B, Bijnens M, Uyttersprot K, Nguyen H, Martinez SE, Schols D, Herdewijn P, Pannecouque C, Arnold E, Das K
PNAS (2022) 119 pp. e2203660119-e2203660119 [ Pubmed: 35858448 DOI: doi:10.1073/pnas.2203660119 ]
- Nnrti resistant m184i/e138k mutant hiv-1 reverse transcriptase with a dna aptamer in complex with rilpivirine (Complex from Human immunodeficiency virus type 1 BH10)
- Dna (38-mer) (11 kDa, DNA from synthetic construct)
- Hiv-1 reverse transcriptase p51 subunit with e138k mutation (Complex from Human immunodeficiency virus type 1 BH10)
- 4-{[4-({4-[(e)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile (366 Da, Ligand)
- Hiv-1 reverse transcriptase p66 subunit with m184i mutation (Complex from Human immunodeficiency virus type 1 BH10)
- P51 rt (50 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (64 kDa, Protein from Human immunodeficiency virus type 1 BH10)
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine
Resolution: 3.65 Å
EM Method: Single-particle
Fitted PDBs: 7z2g
Q-score: 0.466
Singh AK, De Wijngaert B, Bijnens M, Uyttersprot K, Nguyen H, Martinez SE, Schols D, Herdewijn P, Pannecouque C, Arnold E, Das K
PNAS (2022) 119 pp. e2203660119-e2203660119 [ Pubmed: 35858448 DOI: doi:10.1073/pnas.2203660119 ]
- Hiv-1 reverse transcriptase p51 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile (425 Da, Ligand)
- Hiv-1 reverse transcriptase p66 subunit (Complex from Human immunodeficiency virus type 1 BH10)
- Hiv-1 reverse transcriptase with a dna aptamer in complex with doravirine (Complex from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (64 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Reverse transcriptase/ribonuclease h (50 kDa, Protein from Human immunodeficiency virus type 1 BH10)
- Dna (38-mer) (11 kDa, DNA from synthetic construct)