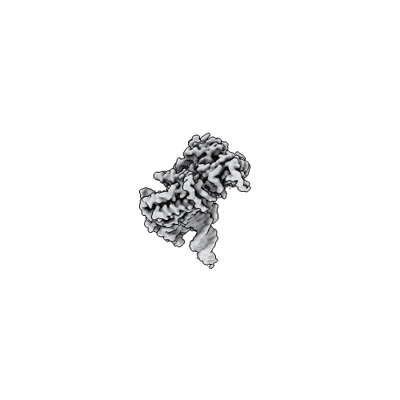
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8ucu
Q-score: 0.52
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)
- Rna-dna primer (9 kDa, DNA from synthetic construct)
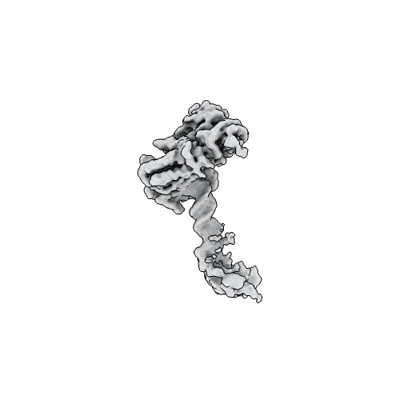
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.81 Å
EM Method: Single-particle
Fitted PDBs: 8ucv
Q-score: 0.337
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Rna-dna/dna primer (9 kDa, Macromolecule from synthetic construct)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)
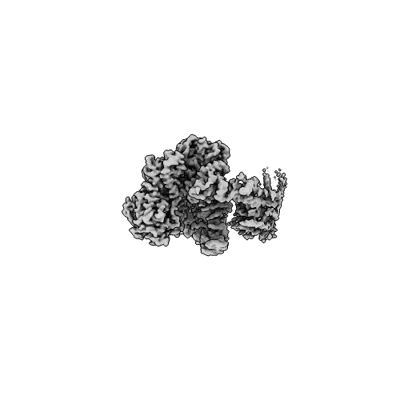
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.64 Å
EM Method: Single-particle
Fitted PDBs: 8ucw
Q-score: 0.377
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Rna-dna/dna primer (9 kDa, Macromolecule from synthetic construct)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)
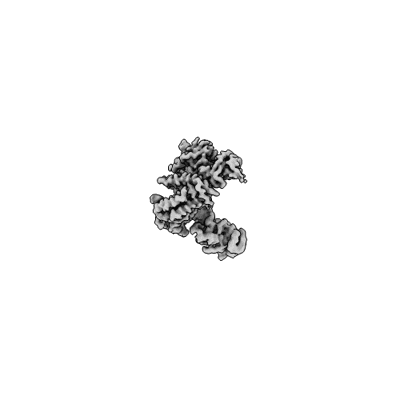
Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8g99
Q-score: 0.551
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- Polymerase alpha-primase (110 kDa, Complex)
- Iron/sulfur cluster (351 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8g9f
Q-score: 0.48
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- Polymerase alpha-primase (110 kDa, Complex)
- Dna primase (49 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8g9l
Q-score: 0.453
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Rna primer (2 kDa, RNA from synthetic construct)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- Polymerase alpha-primase with a dna initiation substrate (110 kDa, Complex)
- Dna initiation substrate (20 kDa, Complex from synthetic construct)
Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8g9n
Q-score: 0.415
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Rna-dna primer (6 kDa, DNA from synthetic construct)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- Polymerase alpha-primase with a dna elongation substrate (20 kDa, Complex)
- Dna elongation substrate (Complex from synthetic construct)
Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 8g9o
Q-score: 0.275
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- Polymerase alpha-primase with a dna elongation substrate (110 kDa, Complex)
- Dna elongation substrate (20 kDa, Complex from synthetic construct)
- Rna-dna primer (6 kDa, Macromolecule from synthetic construct)
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-33 Antibody
Williamson LE, Gilliland Jr T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox JM, Diamond MS, Ohi MD, Klimstra WB, Crowe Jr JE
Cell (2020) 183 pp. 1884-1900.e23 [ Pubmed: 33301709 DOI: doi:10.1016/j.cell.2020.11.011 ]