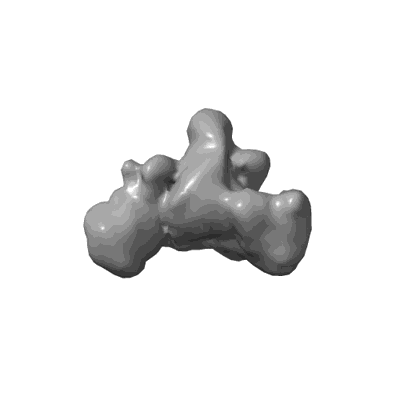
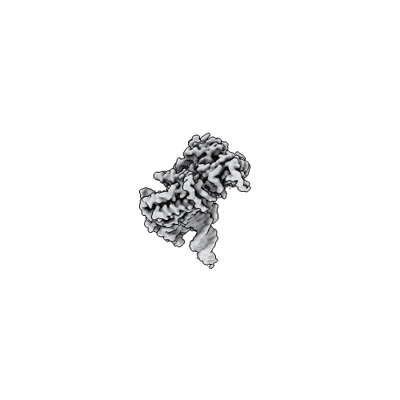
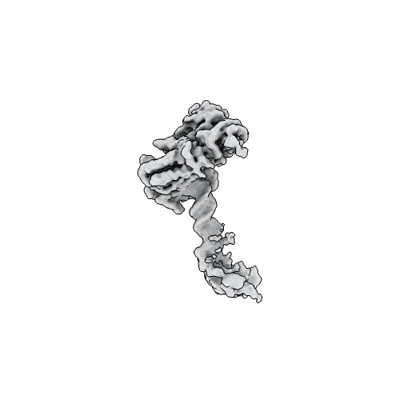
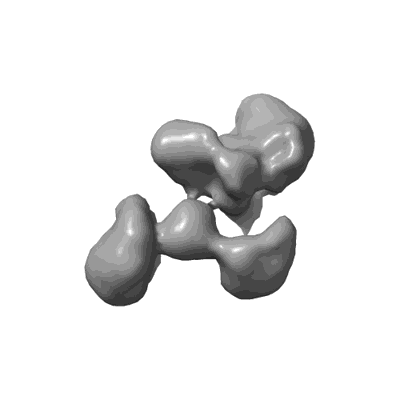Cholinephosphotransferase in complex with CDP-choline and phosphatidylcholine
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8urp
Q-score: 0.589
Roberts JR, Horibata Y, Kwarcinski FE, Lam V, Raczkowski AM, Hubbard A, White B, Sugimoto H, Tall GG, Ohi MD, Maeda S
Nat Commun (2025) 16 pp. 111-111 [ Pubmed: 39747155 DOI: doi:10.1038/s41467-024-55673-1 ]
- Nanobody25 (Complex from Lama glama)
- Cholinephosphotransferase 1 (44 kDa, Protein from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- [2-cytidylate-o'-phosphonyloxyl]-ethyl-trimethyl-ammonium (488 Da, Ligand)
- Ycpt1 (Complex from Saccharomyces cerevisiae)
- Water (18 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (787 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phoshocholine (734 Da, Ligand)
Cholinephosphotransferase in complex with selective inhibitor chelerythrine
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8urt
Q-score: 0.546
Roberts JR, Horibata Y, Kwarcinski FE, Lam V, Raczkowski AM, Hubbard A, White B, Sugimoto H, Tall GG, Ohi MD, Maeda S
Nat Commun (2025) 16 pp. 111-111 [ Pubmed: 39747155 DOI: doi:10.1038/s41467-024-55673-1 ]
- Nanobody25 (Complex from Lama glama)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Ycpt1 (Complex from Saccharomyces cerevisiae)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (787 Da, Ligand)
- 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium (348 Da, Ligand)
- Cholinephosphotransferase 1 (44 kDa, Protein from Saccharomyces cerevisiae)
Cholinephosphotransferase in complex with diacylglycerol
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8ul9
Q-score: 0.503
Roberts JR, Horibata Y, Kwarcinski FE, Lam V, Raczkowski AM, Hubbard A, White B, Sugimoto H, Tall GG, Ohi MD, Maeda S
Nat Commun (2025) 16 pp. 111-111 [ Pubmed: 39747155 DOI: doi:10.1038/s41467-024-55673-1 ]
- Nanobody24 (Complex from Lama glama)
- Cholinephosphotransferase 1 (44 kDa, Protein from Saccharomyces cerevisiae)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Ycpt1 (Complex from Saccharomyces cerevisiae)
- Diacyl glycerol (625 Da, Ligand)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (787 Da, Ligand)
- Saccharomyces cerevisiae cdp-choline phosphotransferase in complex with diacyl-glycerol (Complex from Saccharomyces cerevisiae)
DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Resolution: 11.16 Å
EM Method: Single-particle
Fitted PDBs: 8v6g
Q-score: 0.054
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Rna primer (2 kDa, RNA from synthetic construct)
- Dna primase (49 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Polymerase alpha-primase with a dna initiation substrate (110 kDa, Complex)
- Dna initiation substrate (20 kDa, Complex from synthetic construct)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Resolution: 11.11 Å
EM Method: Single-particle
Fitted PDBs: 8v6h
Q-score: 0.018
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Rna primer (2 kDa, RNA from synthetic construct)
- Dna primase (49 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Polymerase alpha-primase with a dna initiation substrate (110 kDa, Complex)
- Dna initiation substrate (20 kDa, Complex from synthetic construct)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Resolution: 14.06 Å
EM Method: Single-particle
Fitted PDBs: 8v6i
Q-score: 0.032
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Dna primase (49 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Polymerase alpha-primase with a dna elongation substrate (110 kDa, Complex)
- Dna elongation substrate (20 kDa, Complex from synthetic construct)
- Rna-dna primer (6 kDa, Macromolecule from synthetic construct)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Resolution: 11.11 Å
EM Method: Single-particle
Fitted PDBs: 8v6j
Q-score: 0.032
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Primase (Complex from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna template (15 kDa, DNA from synthetic construct)
- Dna primase (49 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Polymerase alpha (Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Polymerase alpha-primase with a dna elongation substrate (110 kDa, Complex)
- Dna elongation substrate (20 kDa, Complex from synthetic construct)
- Rna-dna primer (6 kDa, Macromolecule from synthetic construct)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna polymerase alpha subunit b (67 kDa, Protein from Xenopus laevis)
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase
Resolution: 2.85 Å
EM Method: Single-particle
Fitted PDBs: 8ucu
Q-score: 0.52
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)
- Rna-dna primer (9 kDa, DNA from synthetic construct)
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.81 Å
EM Method: Single-particle
Fitted PDBs: 8ucv
Q-score: 0.337
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Dna primase large subunit (59 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Rna-dna/dna primer (9 kDa, Macromolecule from synthetic construct)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase
Resolution: 3.64 Å
EM Method: Single-particle
Fitted PDBs: 8ucw
Q-score: 0.377
Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF
Nat Struct Mol Biol (2024) 31 pp. 777-790 [ Pubmed: 38491139 DOI: doi:10.1038/s41594-024-01227-4 ]
- Dna polymerase alpha catalytic subunit (129 kDa, Protein from Xenopus laevis)
- Magnesium ion (24 Da, Ligand)
- Dna termination substrate (30 kDa, Complex from synthetic construct)
- Polymerase alpha (200 kDa, Complex from Xenopus laevis)
- 2'-deoxyguanosine-5'-triphosphate (507 Da, Ligand)
- Dna template (18 kDa, DNA from synthetic construct)
- Primase (110 kDa, Complex from Xenopus laevis)
- Rna-dna/dna primer (9 kDa, Macromolecule from synthetic construct)
- Polymerase alpha-primase with a dna termination substrate (330 kDa, Complex)