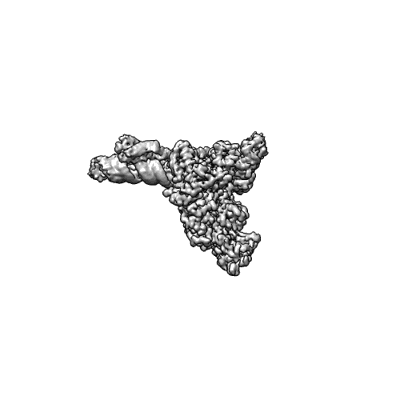
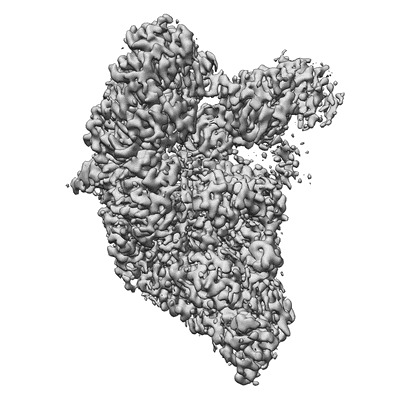
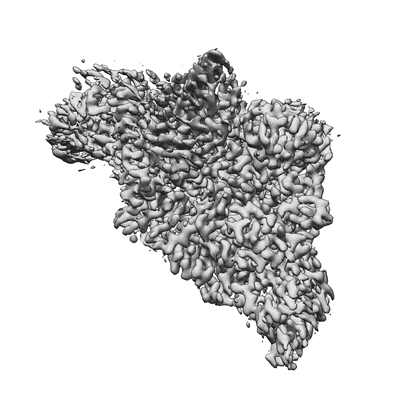
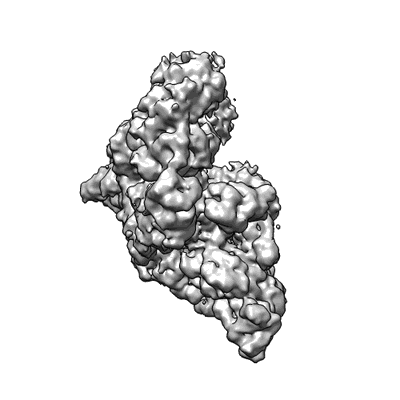
Resolution: 3.38 Å
EM Method: Single-particle
Fitted PDBs: 7uo4
Q-score: 0.457
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Sars-cov-2 replication-transcription complex (Complex from Severe acute respiratory syndrome coronavirus 2)
- [[(2~{r},3~{s},4~{r},5~{r})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate (531 Da, Ligand)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state
Resolution: 3.09 Å
EM Method: Single-particle
Fitted PDBs: 7uo7
Q-score: 0.451
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Sars-cov-2 replication-transcription complex + atp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state.
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7uo9
Q-score: 0.487
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Uridine 5'-triphosphate (484 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 replication-transcription complex + utp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 7uob
Q-score: 0.548
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Zinc ion (65 Da, Ligand)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 replication-transcription complex + gtp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- 3'-deoxyuridine-5'-monophosphate (308 Da, Ligand)
- Water (18 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state
Resolution: 2.67 Å
EM Method: Single-particle
Fitted PDBs: 7uoe
Q-score: 0.523
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Zinc ion (65 Da, Ligand)
- 3'-deoxyuridine-5'-monophosphate (308 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Cytidine-5'-triphosphate (483 Da, Ligand)
- Water (18 Da, Ligand)
- Sars-cov-2 replication-transcription complex + ctp (Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7krn
Q-score: 0.476
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ Pubmed: 33883267 DOI: doi:10.1073/pnas.2102516118 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Aluminum fluoride (83 Da, Ligand)
- Helicase (67 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Sars-cov-2 backtracked complex bound to nsp13 helicase - nsp13(1)-btc (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Chapso (631 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (43-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7kro
Q-score: 0.416
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ Pubmed: 33883267 DOI: doi:10.1073/pnas.2102516118 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Aluminum fluoride (83 Da, Ligand)
- Helicase (67 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-btc (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Chapso (631 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (43-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7krp
Q-score: 0.361
Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, Shaw DE, Landick R, Darst SA, Campbell EA
PNAS (2021) 118 [ Pubmed: 33883267 DOI: doi:10.1073/pnas.2102516118 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 backtracked complex complex bound to nsp13 helicase - btc (local refinement) (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (37-mer) (12 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna (36-mer) (17 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Chapso (631 Da, Ligand)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 replication/transcription complex bound to nsp13 helicase - nsp13(1)-RTC
Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, Maruthi K, Eng ET, Vatandaslar H, Chait B, Kapoor T, Darst SA, Campbell EA
bioRxiv (2020) [ Pubmed: 32676607 DOI: doi:10.1101/2020.07.08.194084 ]
Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, Maruthi K, Eng ET, Vatandaslar H, Chait B, Kapoor T, Darst SA, Campbell EA
bioRxiv (2020) [ Pubmed: 32676607 DOI: doi:10.1101/2020.07.08.194084 ]