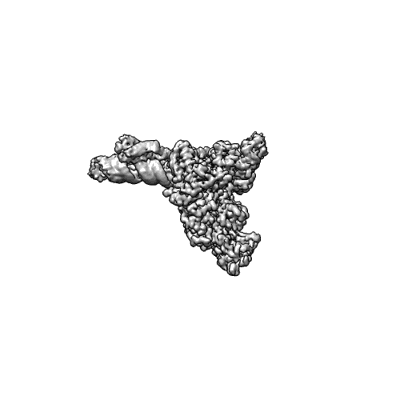
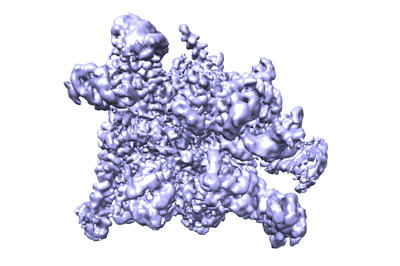
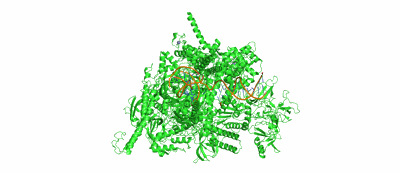
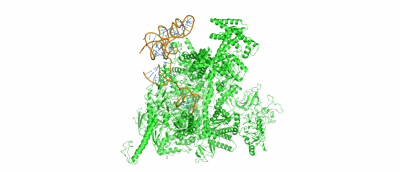
Resolution: 3.38 Å
EM Method: Single-particle
Fitted PDBs: 7uo4
Q-score: 0.457
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Sars-cov-2 replication-transcription complex (Complex from Severe acute respiratory syndrome coronavirus 2)
- [[(2~{r},3~{s},4~{r},5~{r})-5-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate (531 Da, Ligand)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state
Resolution: 3.09 Å
EM Method: Single-particle
Fitted PDBs: 7uo7
Q-score: 0.451
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Sars-cov-2 replication-transcription complex + atp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state.
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7uo9
Q-score: 0.487
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Uridine 5'-triphosphate (484 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 replication-transcription complex + utp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 7uob
Q-score: 0.548
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Zinc ion (65 Da, Ligand)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 replication-transcription complex + gtp (Complex from Severe acute respiratory syndrome coronavirus 2)
- Guanosine-5'-triphosphate (523 Da, Ligand)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- 3'-deoxyuridine-5'-monophosphate (308 Da, Ligand)
- Water (18 Da, Ligand)
SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state
Resolution: 2.67 Å
EM Method: Single-particle
Fitted PDBs: 7uoe
Q-score: 0.523
Malone BF, Perry JK, Olinares PDB, Lee HW, Chen J, Appleby TC, Feng JY, Bilello JP, Ng H, Sotiris J, Ebrahim M, Chua EYD, Mendez JH, Eng ET, Landick R, Gotte M, Chait BT, Campbell EA, Darst SA
Nature (2023) 614 pp. 781-787 [ DOI: doi:10.1038/s41586-022-05664-3 Pubmed: 36725929 ]
- Zinc ion (65 Da, Ligand)
- 3'-deoxyuridine-5'-monophosphate (308 Da, Ligand)
- Rna-directed rna polymerase (106 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Template rna (55-mer) (17 kDa, RNA from synthetic construct)
- Product rna (35-mer) (11 kDa, RNA from synthetic construct)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Cytidine-5'-triphosphate (483 Da, Ligand)
- Water (18 Da, Ligand)
- Sars-cov-2 replication-transcription complex + ctp (Complex from Severe acute respiratory syndrome coronavirus 2)
cryo-EM structure of HK022 putRNA-associated E.coli RNA polymerase elongation complex
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 7xue
Q-score: 0.476
Hwang S, Olinares PDB, Lee J, Kim J, Chait BT, King RA, Kang JY
Nat Commun (2022) 13 pp. 4668-4668 [ DOI: doi:10.1038/s41467-022-32315-y Pubmed: 35970830 ]
- Zinc ion (65 Da, Ligand)
- Rna (nun gene and immunity region) (30 kDa, RNA from Shamshuipovirus HK022)
- Dna/rna (Complex)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit beta' (158 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli (strain K12))
- Hk022 putrna-associated e.coli rna polymerase elongation complex (440 kDa, Complex)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli (strain K12))
- Magnesium ion (24 Da, Ligand)
- Template dna (54 kDa, DNA from Shamshuipovirus HK022)
- Rna polymerase (Complex from Escherichia coli (strain K12))
- Non-template dna (54 kDa, DNA from Shamshuipovirus HK022)
cryo-EM structure of HK022 putRNA-less E.coli RNA polymerase elongation complex
Resolution: 3.57 Å
EM Method: Single-particle
Fitted PDBs: 7xug
Q-score: 0.446
Hwang S, Olinares PDB, Lee J, Kim J, Chait BT, King RA, Kang JY
Nat Commun (2022) 13 pp. 4668-4668 [ DOI: doi:10.1038/s41467-022-32315-y Pubmed: 35970830 ]
- Zinc ion (65 Da, Ligand)
- Rna (nun gene and immunity region) (30 kDa, RNA from Shamshuipovirus HK022)
- Dna/rna (Complex)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli (strain K12))
- Hk022 putrna-less e.coli rna polymerase elongation complex (440 kDa, Complex)
- Dna-directed rna polymerase subunit beta' (158 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli (strain K12))
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli (strain K12))
- E.coli rna polymerase (Complex from Escherichia coli (strain K12))
- Magnesium ion (24 Da, Ligand)
- Template dna (54 kDa, DNA from Shamshuipovirus HK022)
- Non-template dna (54 kDa, DNA from Shamshuipovirus HK022)
Cryo-EM structure of sigma70 bound HK022 putRNA-associated E.coli RNA polymerase elongation complex
Resolution: 3.61 Å
EM Method: Single-particle
Fitted PDBs: 7xui
Q-score: 0.416
Hwang S, Olinares PDB, Lee J, Kim J, Chait BT, King RA, Kang JY
Nat Commun (2022) 13 pp. 4668-4668 [ DOI: doi:10.1038/s41467-022-32315-y Pubmed: 35970830 ]
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli K-12)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli K-12)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli K-12)
- Zinc ion (65 Da, Ligand)
- Dna/rna (Complex)
- Rna (122-mer) (39 kDa, RNA from Shamshuipovirus HK022)
- Sigma70 bound hk022 putrna-associated e.coli rna polymerase elongation complex (510 kDa, Complex)
- Dna-directed rna polymerase subunit beta' (158 kDa, Protein from Escherichia coli K-12)
- Magnesium ion (24 Da, Ligand)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli K-12)
- Template dna (54 kDa, DNA from Shamshuipovirus HK022)
- Rna polymerase (Complex from Escherichia coli (strain K12))
Mfd-bound E.coli RNA polymerase elongation complex - L1 state
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6x26
Q-score: 0.339
Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, Darst SA
eLife (2021) 10 [ DOI: doi:10.7554/eLife.62117 Pubmed: 33480355 ]
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli)
- Rna (20-mer) (6 kDa, RNA from Escherichia coli)
- Transcription-repair-coupling factor (130 kDa, Protein from Escherichia coli)
- Escherichia coli mfd-rnap elonation complex: state l1 (570 kDa, Complex from Escherichia coli)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli)
- Dna (64-mer) (19 kDa, DNA from Escherichia coli)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna (64-mer) (19 kDa, DNA from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli)
Mfd-bound E.coli RNA polymerase elongation complex - L2 state
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6x2f
Q-score: 0.341
Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, Darst SA
eLife (2021) 10 [ DOI: doi:10.7554/eLife.62117 Pubmed: 33480355 ]
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli)
- Transcription-repair-coupling factor (130 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli)
- Rna (20-mer) (6 kDa, RNA from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Dna (64-mer) (19 kDa, DNA from Escherichia coli)
- Escherichia coli mfd-rnap elongation complex: l2 state (570 kDa, Complex from Escherichia coli)
- Dna (64-mer) (19 kDa, DNA from Escherichia coli)