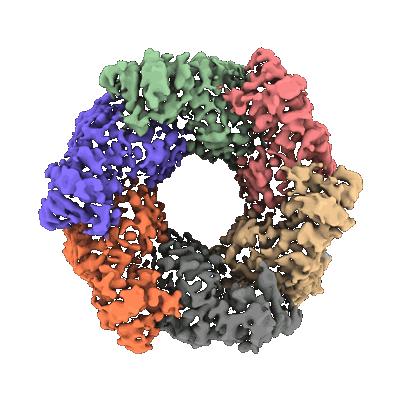
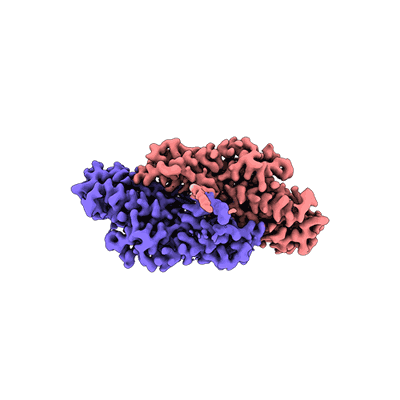
Resolution: 4.16 Å
EM Method: Single-particle
Fitted PDBs: 9bei
Q-score: 0.276
Goverde CA, Pacesa M, Goldbach N, Dornfeld LJ, Balbi PEM, Georgeon S, Rosset S, Kapoor S, Choudhury J, Dauparas J, Schellhaas C, Kozlov S, Baker D, Ovchinnikov S, Vecchio AJ, Correia BE
Nature (2024) 631 pp. 449-458 [ DOI: doi:10.1038/s41586-024-07601-y Pubmed: 38898281 ]
- Cop-2 fab light chain (23 kDa, Protein from Escherichia coli)
- Claudin-4 (22 kDa, Protein from Homo sapiens)
- Synthetic human claudin-4 complex with clostridium perfringens enterotoxin c-terminal domain, sfab cop-2, and nanobody against cop-2 (103 kDa, Complex from Homo sapiens)
- Cop-2 fab heavy chain (25 kDa, Protein from Escherichia coli)
- Heat-labile enterotoxin b chain (14 kDa, Protein from Clostridium perfringens)
- Anti-fab nanobody (13 kDa, Protein from Escherichia coli)
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus
Resolution: 2.79 Å
EM Method: Single-particle
Fitted PDBs: 8p49
Q-score: 0.504
Schweke H, Pacesa M, Levin T, Goverde CA, Kumar P, Duhoo Y, Dornfeld LJ, Dubreuil B, Georgeon S, Ovchinnikov S, Woolfson DN, Correia BE, Dey S, Levy ED
Cell (2024) 187 pp. 999- [ DOI: doi:10.1016/j.cell.2024.01.022 Pubmed: 38325366 ]
Cysteine tRNA ligase homodimer
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8qhp
Q-score: 0.546
Schweke H, Pacesa M, Levin T, Goverde CA, Kumar P, Duhoo Y, Dornfeld LJ, Dubreuil B, Georgeon S, Ovchinnikov S, Woolfson DN, Correia BE, Dey S, Levy ED
Cell (2024) 187 pp. 999- [ DOI: doi:10.1016/j.cell.2024.01.022 Pubmed: 38325366 ]
- Homodimeric form of cysteine trna ligase (114 kDa, Complex from Pyrococcus furiosus DSM 3638)
- Cysteine--trna ligase (57 kDa, Protein from Pyrococcus furiosus DSM 3638)
- Zinc ion (65 Da, Ligand)
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 8sk7
Q-score: 0.528
Watson JL, Juergens D, Bennett NR, Trippe BL, Yim J, Eisenach HE, Ahern W, Borst AJ, Ragotte RJ, Milles LF, Wicky BIM, Hanikel N, Pellock SJ, Courbet A, Sheffler W, Wang J, Venkatesh P, Sappington I, Torres SV, Lauko A, De Bortoli V, Mathieu E, Ovchinnikov S, Barzilay R, Jaakkola TS, DiMaio F, Baek M, Baker D
Nature (2023) 620 pp. 1089-1100 [ Pubmed: 37433327 DOI: doi:10.1038/s41586-023-06415-8 ]
- Influenza ha (iowa43) bound to rfdiffusion designed minibinder, ha_20 (Complex from Influenza A virus)
- Hemagglutinin (26 kDa, Protein from Influenza A virus)
- Hemagglutinin ha1 chain (35 kDa, Protein from Influenza A virus)
- Ha_20 minibinder (rfdiffusion-designed) (7 kDa, Protein from synthetic construct)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7abr
Q-score: 0.454
Morreale FE, Kleine S, Leodolter J, Junker S, Hoi DM, Ovchinnikov S, Okun A, Kley J, Kurzbauer R, Junk L, Guha S, Podlesainski D, Kazmaier U, Boehmelt G, Weinstabl H, Rumpel K, Schmiedel VM, Hartl M, Haselbach D, Meinhart A, Kaiser M, Clausen T
Cell (2022) 185 pp. 2338- [ Pubmed: 35662409 DOI: doi:10.1016/j.cell.2022.05.009 ]
- Negative regulator of genetic competence clpc/mecb (91 kDa, Protein from Bacillus subtilis (strain 168))
- Negative regulator of genetic competence clpc/mecb (Complex from Bacillus subtilis subsp. subtilis str. 168)
- Substrate polypeptide (Complex from unidentified)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- B. subtilis clpc (dwb mutant) hexamer bound to a substrate polypeptide (Complex from Bacillus subtilis subsp. subtilis str. 168)
- Substrate polypeptide (2 kDa, Protein from unidentified)
Cryo-EM map of Hrd1/Hrd3 monomer
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6vjy
Q-score: 0.334
Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA
Science (2020) 368 [ Pubmed: 32327568 DOI: doi:10.1126/science.aaz2449 ]
CryoEM map of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6vjz
Q-score: 0.334
Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA
Science (2020) 368 [ Pubmed: 32327568 DOI: doi:10.1126/science.aaz2449 ]
- Erad-associated e3 ubiquitin-protein ligase component hrd3 (88 kDa, Protein from Saccharomyces cerevisiae)
- Complex of hrd1-usa1/der1/hrd3 in the expected topology (Complex from Saccharomyces cerevisiae)
- U1 snp1-associating protein 1 (39 kDa, Protein from Saccharomyces cerevisiae)
- Degradation in the endoplasmic reticulum protein 1 (24 kDa, Protein from Saccharomyces cerevisiae)
- Erad-associated e3 ubiquitin-protein ligase hrd1 (55 kDa, Protein from Saccharomyces cerevisiae)
CryoEM map of Hrd1-Usa1/Der1/Hrd3 of the flipped topology
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6vk0
Q-score: 0.369
Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA
Science (2020) 368 [ Pubmed: 32327568 DOI: doi:10.1126/science.aaz2449 ]
- Erad-associated e3 ubiquitin-protein ligase component hrd3 (88 kDa, Protein from Saccharomyces cerevisiae)
- U1 snp1-associating protein 1 (39 kDa, Protein from Saccharomyces cerevisiae)
- Complex of hrd1-usa1/der1/hrd3 of the flipped topology (Complex from Saccharomyces cerevisiae)
- Degradation in the endoplasmic reticulum protein 1 (24 kDa, Protein from Saccharomyces cerevisiae)
- Erad-associated e3 ubiquitin-protein ligase hrd1 (55 kDa, Protein from Saccharomyces cerevisiae)
CryoEM map of Hrd1/Hrd3 part from Hrd1-Usa1/Der1/Hrd3 complex
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6vk1
Q-score: 0.401
Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA
Science (2020) 368 [ Pubmed: 32327568 DOI: doi:10.1126/science.aaz2449 ]
CryoEM map of Hrd3/Yos9 complex
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 6vk3
Q-score: 0.405
Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA
Science (2020) 368 [ Pubmed: 32327568 DOI: doi:10.1126/science.aaz2449 ]
- Hrd3 (84 kDa, Protein from Saccharomyces cerevisiae)
- A complex of hrd1/hrd3/yos9 (Complex from Saccharomyces cerevisiae)
- Protein os-9 homolog (61 kDa, Protein from Saccharomyces cerevisiae)