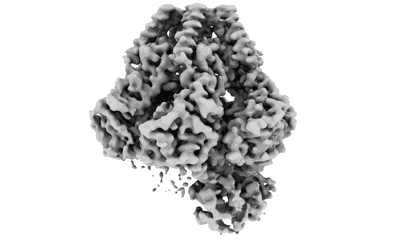
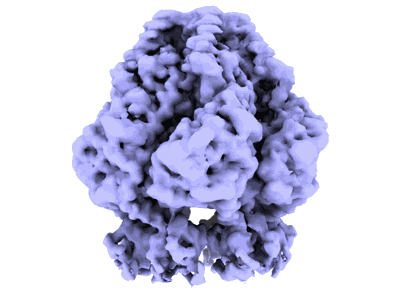
Trimer of the dimeric SaPI2 Stl transcriptional regulator
Debiasi-Anders G, Qiao C, Salim A, Li N, Mir-Sanchis I
Nat Commun (2025) 16 pp. 1889-1889 [ Pubmed: 39987160 DOI: doi:10.1038/s41467-025-57156-3 ]
- Helix-turn-helix xre family protein (27 kDa, Protein from Staphylococcus aureus)
- Water (18 Da, Ligand)
- Trimer of the dimeric sapi2 stl transcriptional regulator (600 kDa, Cellular component from Staphylococcus aureus)
Complex between the 80a-Sak SSAP and the SaPI2 Stl master regulator
Debiasi-Anders G, Qiao C, Salim A, Li N, Mir-Sanchis I
Nat Commun (2025) 16 pp. 1889-1889 [ Pubmed: 39987160 DOI: doi:10.1038/s41467-025-57156-3 ]
- Helix-turn-helix xre family protein (27 kDa, Protein from Staphylococcus aureus)
- Duf1071 domain-containing protein (23 kDa, Protein from Staphylococcus phage 80alpha)
- 80a-sak ssap (Complex from Staphylococcus phage 80alpha)
- Sapi2 stl master regulator (Complex from Staphylococcus aureus)
- Complex between the 80a-sak ssap and the sapi2 stl master regulator (1 MDa, Complex)
Complex between the RecA-like Sak4 SSAP and the SaPI2 Stl master regulator
Debiasi-Anders G, Qiao C, Salim A, Li N, Mir-Sanchis I
Nat Commun (2025) 16 pp. 1889-1889 [ Pubmed: 39987160 DOI: doi:10.1038/s41467-025-57156-3 ]
- Complex between sak4 ssap from staphylococcal phage 52a and the sapi2 stl master regulator (Complex from Staphylococcus aureus)
- Helix-turn-helix xre family protein (27 kDa, Protein from Staphylococcus aureus)
- Orf016 (31 kDa, Protein from Staphylococcus phage 52A)
- Phosphothiophosphoric acid-adenylate ester (523 Da, Ligand)
Sak Single Strand Annealing Protein from Staphylococcal Bacteriophage 80a - dCTD
Debiasi-Anders G, Qiao C, Salim A, Li N, Mir-Sanchis I
Nat Commun (2025) 16 pp. 1889-1889 [ Pubmed: 39987160 DOI: doi:10.1038/s41467-025-57156-3 ]
- 18-mer ring cryoem structure of the 80a-sak ntd in apo state (460 kDa, Complex from Staphylococcus phage 80alpha)
- Water (18 Da, Ligand)
- Topoisomerase (19 kDa, Protein from Staphylococcus phage 80alpha)
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) [ Pubmed: 35871290 DOI: 10.1093/nar/gkac625 ]
Image sets:
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) 50 pp. 8349-8362 [ Pubmed: 35871290 DOI: doi:10.1093/nar/gkac625 ]
- Rep1 in sapi1 (330 kDa, Cellular component from Staphylococcus aureus)
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) [ Pubmed: 35871290 DOI: 10.1093/nar/gkac625 ]
Image sets:
Structure of Primase-Helicase in SaPI5
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7ola
Q-score: 0.449
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) 50 pp. 8349-8362 [ Pubmed: 35871290 DOI: doi:10.1093/nar/gkac625 ]
- Primase-helicase in sapi5 (669 kDa, Cellular component from Staphylococcus aureus)
- Dna primase (93 kDa, Protein from Staphylococcus aureus)
- Magnesium ion (24 Da, Ligand)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
Structure of Primase-Helicase in SaPI5
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7om0
Q-score: 0.441
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) 50 pp. 8349-8362 [ Pubmed: 35871290 DOI: doi:10.1093/nar/gkac625 ]
- Primase-helicase in sapi5 (669 kDa, Cellular component from Staphylococcus aureus)
- Dna primase (93 kDa, Protein from Staphylococcus aureus)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
Structure of Rep protein in SaPI1
Qiao C, Debiasi-Anders G, Mir-Sanchis I
Nucleic Acids Res (2022) 50 pp. 8349-8362 [ Pubmed: 35871290 DOI: doi:10.1093/nar/gkac625 ]
- Rep1 in sapi1 (330 kDa, Cellular component from Staphylococcus aureus)
- Rep1 (Protein from Staphylococcus aureus)