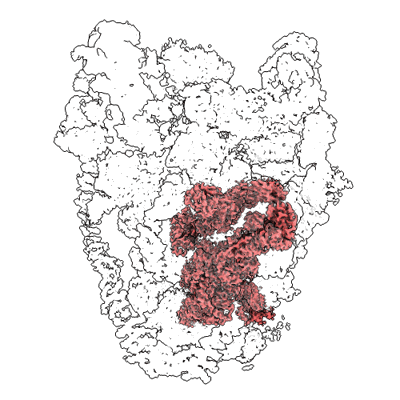
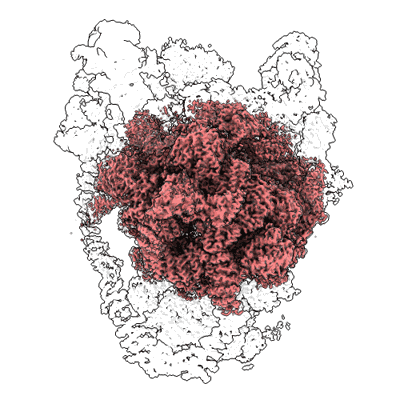
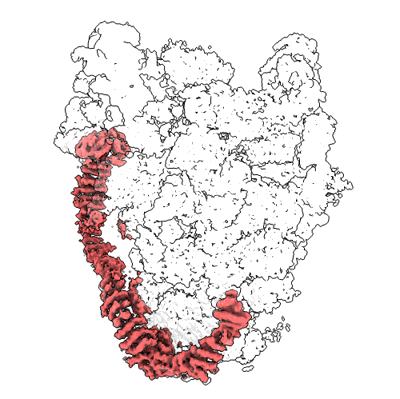
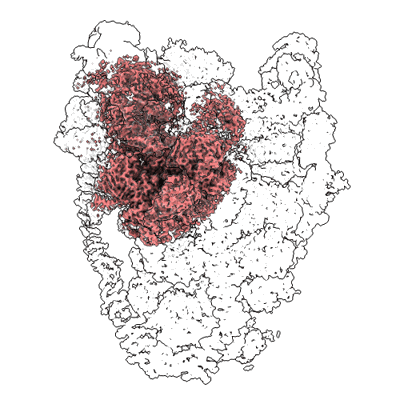
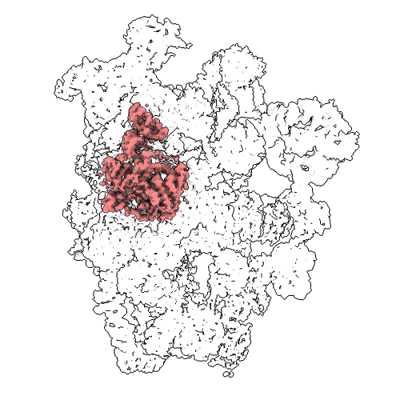
Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate
Resolution: 3.13 Å
EM Method: Single-particle
Fitted PDBs: 7vyq
Q-score: 0.578
Li Y, Zhang R, Wang C, Forouhar F, Clarke OB, Vorobiev S, Singh S, Montelione GT, Szyperski T, Xu Y, Hunt JF
EMBO J (2022) 41 pp. e108368-e108368 [ Pubmed: 35801308 DOI: doi:10.15252/embj.2021108368 ]
- Carbonyl reductase (30 kDa, Protein from Candida parapsilosis)
- Ternary complex of scr with nadp and ethyl 4-chloroacetoacetate (Complex from Candida parapsilosis)
- Nadp nicotinamide-adenine-dinucleotide phosphate (743 Da, Ligand)
- Ethyl 4-chloranyl-3-oxidanylidene-butanoate (164 Da, Ligand)
Oligomeric interactions maintain active-site structure in a non-cooperative enzyme family
Li Y, Zhang R, Wang C, Forouhar F, Clarke OB, Vorobiev S, Singh S, Montelione GT, Szyperski T, Xu Y, Hunt JF
EMBO J (2022) pp. e108368-e108368 [ Pubmed: 35801308 DOI: doi:10.15252/embj.2021108368 ]
Oligomeric interactions maintain active-site structure in a non-cooperative enzyme family
Li Y, Zhang R, Wang C, Forouhar F, Clarke OB, Vorobiev S, Singh S, Montelione GT, Szyperski T, Xu Y, Hunt JF
EMBO J (2022) pp. e108368-e108368 [ Pubmed: 35801308 DOI: doi:10.15252/embj.2021108368 ]
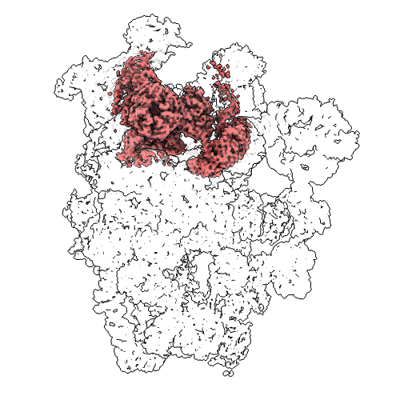
Cryo-EM structure of the human SSU processome, state pre-A1 - 5'ETS focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
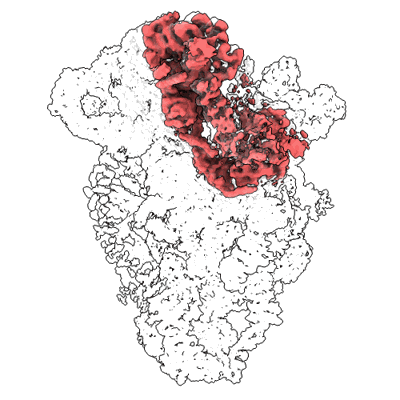
Cryo-EM structure of the human SSU processome, state pre-A1 - Core focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
Cryo-EM structure of the human SSU processome, state pre-A1 - HEATR1 focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
Cryo-EM structure of the human SSU processome, state pre-A1 - NGDN focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
Cryo-EM structure of the human SSU processome, state pre-A1 - PNO1 focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
Cryo-EM structure of the human SSU processome, state pre-A1 - TBL3 focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)
Cryo-EM structure of the human SSU processome, state pre-A1 - UTP20-CTD focused map
Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S
Science (2021) 373 pp. eabj5338-eabj5338 [ Pubmed: 34516797 DOI: doi:10.1126/science.abj5338 ]
- Human ssu processome, state post-a1 (5 MDa, Complex from Homo sapiens)