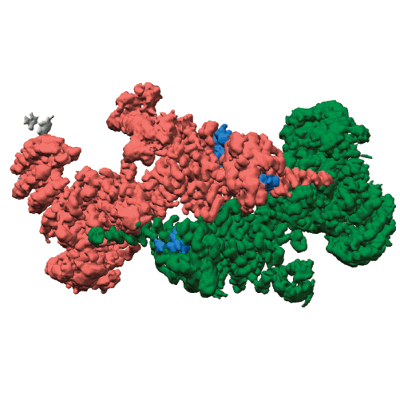
Structure of CUL9-RBX1 ubiquitin E3 ligase complex - hexameric assembly
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 8q7e
Q-score: 0.301
Horn-Ghetko D, Hopf LVM, Tripathi-Giesgen I, Du J, Kostrhon S, Vu DT, Beier V, Steigenberger B, Prabu JR, Stier L, Bruss EM, Mann M, Xiong Y, Schulman BA
Nat Struct Mol Biol (2024) 31 pp. 1083-1094 [ Pubmed: 38605244 DOI: doi:10.1038/s41594-024-01257-y ]
- Cullin-9 (281 kDa, Protein from Homo sapiens)
- E3 ubiquitin-protein ligase rbx1 (12 kDa, Protein from Homo sapiens)
- Structure of cul9-rbx1 ubiquitin e3 ligase complex - hexameric assembly (Complex from Homo sapiens)
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 8q7h
Q-score: 0.39
Horn-Ghetko D, Hopf LVM, Tripathi-Giesgen I, Du J, Kostrhon S, Vu DT, Beier V, Steigenberger B, Prabu JR, Stier L, Bruss EM, Mann M, Xiong Y, Schulman BA
Nat Struct Mol Biol (2024) 31 pp. 1083-1094 [ Pubmed: 38605244 DOI: doi:10.1038/s41594-024-01257-y ]
- Nedd8 (9 kDa, Protein from Homo sapiens)
- Unknown (12 kDa, Protein from Homo sapiens)
- E3 ubiquitin-protein ligase rbx1 (12 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Cullin-9 (281 kDa, Protein from Homo sapiens)
- Structure of cul9-rbx1 ubiquitin e3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer (Complex from Homo sapiens)
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8rhz
Q-score: 0.425
Horn-Ghetko D, Hopf LVM, Tripathi-Giesgen I, Du J, Kostrhon S, Vu DT, Beier V, Steigenberger B, Prabu JR, Stier L, Bruss EM, Mann M, Xiong Y, Schulman BA
Nat Struct Mol Biol (2024) 31 pp. 1083-1094 [ Pubmed: 38605244 DOI: doi:10.1038/s41594-024-01257-y ]
- Cullin-9 (281 kDa, Protein from Homo sapiens)
- Structure of cul9-rbx1 ubiquitin e3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer (Complex from Homo sapiens)
- E3 ubiquitin-protein ligase rbx1 (12 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
cryo-EM structure of Doa10 in MSP1E3D1
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 8pd0
Q-score: 0.449
Botsch JJ, Junker R, Sorgenfrei M, Ogger PP, Stier L, von Gronau S, Murray PJ, Seeger MA, Schulman BA, Brauning B
Nat Commun (2024) 15 pp. 410-410 [ DOI: doi:10.1038/s41467-023-44670-5 Pubmed: 38195637 ]
- Doa10 with ubc6 and sybody in msp1e3d1 (150 kDa, Complex from Saccharomyces cerevisiae)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (787 Da, Ligand)
- 1,2-dipalmitoyl-sn-glycero-3-phosphate (647 Da, Ligand)
- Erad-associated e3 ubiquitin-protein ligase doa10 (151 kDa, Protein from Saccharomyces cerevisiae)
cryo-EM structure of Doa10 with RING domain in MSP1E3D1
Resolution: 3.58 Å
EM Method: Single-particle
Fitted PDBs: 8pda
Q-score: 0.419
Botsch JJ, Junker R, Sorgenfrei M, Ogger PP, Stier L, von Gronau S, Murray PJ, Seeger MA, Schulman BA, Brauning B
Nat Commun (2024) 15 pp. 410-410 [ DOI: doi:10.1038/s41467-023-44670-5 Pubmed: 38195637 ]
- Doa10 with ubc6 and sybody in msp1e3d1 (150 kDa, Complex from Saccharomyces cerevisiae)
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (787 Da, Ligand)
- 1,2-dipalmitoyl-sn-glycero-3-phosphate (647 Da, Ligand)
- Erad-associated e3 ubiquitin-protein ligase doa10 (151 kDa, Protein from Saccharomyces cerevisiae)