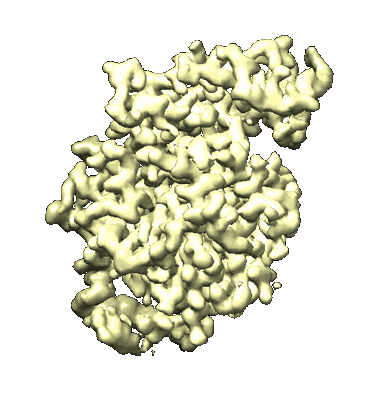
Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae
Resolution: 2.2 Å
EM Method: Single-particle
Fitted PDBs: 8wci
Q-score: 0.725
Suzuki K, Goto Y, Otomo A, Shimizu K, Abe S, Moriyama K, Yasuda S, Hashimoto Y, Kurushima J, Mikuriya S, Imai FL, Adachi N, Kawasaki M, Sato Y, Ogasawara S, Iwata I, Senda T, Ikeguchi M, Tomita H, Iino R, Moriya T, Murata T
To Be Published
- Cardiolipin (1 kDa, Ligand)
- Water (18 Da, Ligand)
- V-type sodium atpase subunit i (76 kDa, Protein from Enterococcus hirae ATCC 9790)
- N,n-dimethyl-4-(5-methyl-1h-benzimidazol-2-yl)aniline (251 Da, Ligand)
- Sodium ion (22 Da, Ligand)
- V-type sodium atpase subunit k (16 kDa, Protein from Enterococcus hirae ATCC 9790)
- V-atpase (735 kDa, Complex from Enterococcus hirae ATCC 9790)
Cryo-EM Structure of inhibitor-free hERG Channel
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8zyn
Q-score: 0.469
Miyashita Y, Moriya T, Kato T, Kawasaki M, Yasuda S, Adachi N, Suzuki K, Ogasawara S, Saito T, Senda T, Murata T
Structure (2024) 32 pp. 1926- [ Pubmed: 39321803 DOI: doi:10.1016/j.str.2024.08.021 ]
- Potassium channel (Complex from Homo sapiens)
- Potassium voltage-gated channel subfamily h member 2 (91 kDa, Protein from Homo sapiens)
Cryo-EM Structure of astemizole-bound hERG Channel
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 8zyo
Q-score: 0.479
Miyashita Y, Moriya T, Kato T, Kawasaki M, Yasuda S, Adachi N, Suzuki K, Ogasawara S, Saito T, Senda T, Murata T
Structure (2024) 32 pp. 1926- [ Pubmed: 39321803 DOI: doi:10.1016/j.str.2024.08.021 ]
Cryo-EM Structure of E-4031-bound hERG Channel
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 8zyp
Q-score: 0.503
Miyashita Y, Moriya T, Kato T, Kawasaki M, Yasuda S, Adachi N, Suzuki K, Ogasawara S, Saito T, Senda T, Murata T
Structure (2024) 32 pp. 1926- [ Pubmed: 39321803 DOI: doi:10.1016/j.str.2024.08.021 ]
Cryo-EM Structure of pimozide-bound hERG Channel
Resolution: 3.18 Å
EM Method: Single-particle
Fitted PDBs: 8zyq
Q-score: 0.488
Miyashita Y, Moriya T, Kato T, Kawasaki M, Yasuda S, Adachi N, Suzuki K, Ogasawara S, Saito T, Senda T, Murata T
Structure (2024) 32 pp. 1926- [ Pubmed: 39321803 DOI: doi:10.1016/j.str.2024.08.021 ]
P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent
Resolution: 2.49 Å
EM Method: Single-particle
Fitted PDBs: 8y6h
Q-score: 0.608
Hamaguchi-Suzuki N, Adachi N, Moriya T, Yasuda S, Kawasaki M, Suzuki K, Ogasawara S, Anzai N, Senda T, Murata T
Biochem Biophys Res Commun (2024) 709 pp. 149855-149855 [ DOI: doi:10.1016/j.bbrc.2024.149855 Pubmed: 38579618 ]
- Uic2 fab heavy chain (24 kDa, Protein from Mus musculus)
- Elacridar (563 Da, Ligand)
- Multidrug resistance protein (Complex from Homo sapiens)
- Atp-dependent translocase abcb1,mneongreen (170 kDa, Protein from Homo sapiens)
- Uic2 fab light chain (24 kDa, Protein from Mus musculus)
P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc
Resolution: 2.54 Å
EM Method: Single-particle
Fitted PDBs: 8y6i
Q-score: 0.576
Hamaguchi-Suzuki N, Adachi N, Moriya T, Yasuda S, Kawasaki M, Suzuki K, Ogasawara S, Anzai N, Senda T, Murata T
Biochem Biophys Res Commun (2024) 709 pp. 149855-149855 [ DOI: doi:10.1016/j.bbrc.2024.149855 Pubmed: 38579618 ]
- Uic2 fab heavy chain (24 kDa, Protein from Mus musculus)
- 1,2-distearoyl-sn-glycerophosphoethanolamine (748 Da, Ligand)
- Elacridar (563 Da, Ligand)
- Multidrug resistance protein (Complex from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- Atp-dependent translocase abcb1,mneongreen (170 kDa, Protein from Homo sapiens)
- Uic2 fab light chain (24 kDa, Protein from Mus musculus)
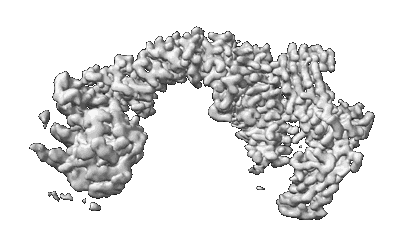
Human VPS29/VPS35L Complex (Locally refined map)
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8sym
Q-score: 0.446
Human VPS35L/VPS29/VPS26C Complex
Resolution: 2.94 Å
EM Method: Single-particle
Fitted PDBs: 8syn
Q-score: 0.38
Boesch DJ, Singla A, Han Y, Kramer DA, Liu Q, Suzuki K, Juneja P, Zhao X, Long X, Medlyn MJ, Billadeau DD, Chen Z, Chen B, Burstein E
Nat Struct Mol Biol (2024) 31 pp. 910-924 [ Pubmed: 38062209 DOI: doi:10.1038/s41594-023-01184-4 ]
- Vacuolar protein sorting-associated protein 26c (33 kDa, Protein from Homo sapiens)
- Trimeric complex of full-length vps35l/vps29/vps26c (Complex from Homo sapiens)
- Vps35 endosomal protein-sorting factor-like (109 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 29 (23 kDa, Protein from Homo sapiens)
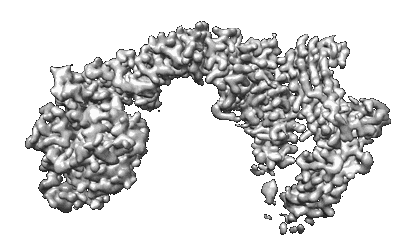
Human Retriever VPS35L/VPS29/VPS26C Complex (Composite Map)
Resolution: 2.94 Å
EM Method: Single-particle
Fitted PDBs: 8syo
Q-score: 0.429
Boesch DJ, Singla A, Han Y, Kramer DA, Liu Q, Suzuki K, Juneja P, Zhao X, Long X, Medlyn MJ, Billadeau DD, Chen Z, Chen B, Burstein E
Nat Struct Mol Biol (2024) 31 pp. 910-924 [ Pubmed: 38062209 DOI: doi:10.1038/s41594-023-01184-4 ]
- Vacuolar protein sorting-associated protein 26c (33 kDa, Protein from Homo sapiens)
- Trimeric complex of full-length vps35l/vps29/vps26c (Complex from Homo sapiens)
- Vps35 endosomal protein-sorting factor-like (109 kDa, Protein from Homo sapiens)
- Vacuolar protein sorting-associated protein 29 (23 kDa, Protein from Homo sapiens)