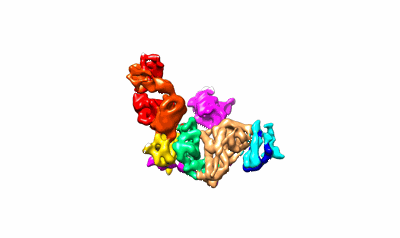
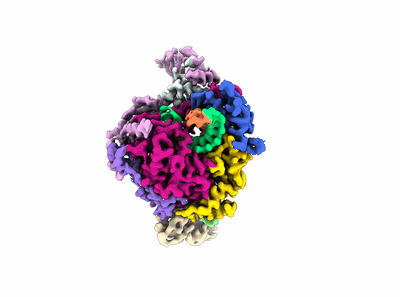Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer
Resolution: 3.74 Å
EM Method: Single-particle
Fitted PDBs: 8tr3
Q-score: 0.426
Wang S, Chan KW, Wei D, Ma X, Liu S, Hu G, Park S, Pan R, Gu Y, Nazzari AF, Olia AS, Xu K, Lin BC, Louder MK, McKee K, Doria-Rose NA, Montefiori D, Seaman MS, Zhou T, Kwong PD, Arthos J, Kong XP, Lu S
Nat Commun (2024) 15 pp. 4301-4301 [ Pubmed: 38773089 DOI: doi:10.1038/s41467-024-48514-8 ]
- Cne40 sosip envelope glycoprotein gp120 (55 kDa, Protein from Human immunodeficiency virus 1)
- Hmab64 fv light chain (11 kDa, Protein from Homo sapiens)
- 3d structure of hmab64 fv in complex with cne40 sosip trimer (Complex)
- Cne40 sosip trimer (Complex from Human immunodeficiency virus 1)
- Cne40 sosip transmembrane protein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Hmab64 fv (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Hmab64 fv heavy chain (13 kDa, Protein from Homo sapiens)
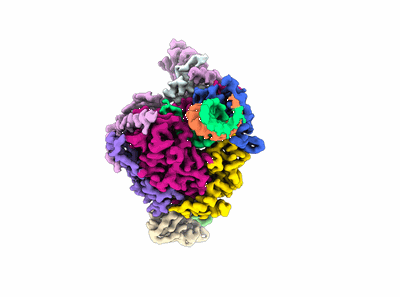
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Resolution: 3.64 Å
EM Method: Single-particle
Fitted PDBs: 7yoy
Q-score: 0.464
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ DOI: doi:10.1016/j.xcrm.2023.101296 Pubmed: 37992686 ]
- 3e8 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Gp42 (Complex from Human gammaherpesvirus 4)
- 5e3,3e8 (Complex from Oryctolagus cuniculus)
- 5e3 heavy chain (13 kDa, Protein from Oryctolagus cuniculus)
- Soluble gp42 (15 kDa, Protein from Human gammaherpesvirus 4)
- 3e8 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- 5e3 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Ebv ghgl-gp42 in complex with mabs 3e8 and 5e3 (Complex)
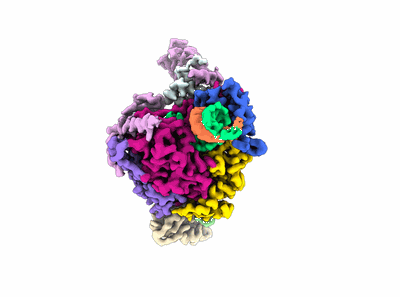
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Resolution: 3.54 Å
EM Method: Single-particle
Fitted PDBs: 7yp1
Q-score: 0.523
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ DOI: doi:10.1016/j.xcrm.2023.101296 Pubmed: 37992686 ]
- Ebv gl (8 kDa, Protein from Human gammaherpesvirus 4)
- 10e4 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Ebv gh (22 kDa, Protein from Human gammaherpesvirus 4)
- Gh,gl (Complex from Human gammaherpesvirus 4)
- 10e4 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- 10e4 (Complex from Oryctolagus cuniculus)
- Ghgl-gp42 in complex with mab 10e4 (Complex)
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ DOI: doi:10.1016/j.xcrm.2023.101296 Pubmed: 37992686 ]
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Resolution: 3.52 Å
EM Method: Single-particle
Fitted PDBs: 7yp2
Q-score: 0.53
Hong J, Zhong L, Liu L, Wu Q, Zhang W, Chen K, Wei D, Sun H, Zhou X, Zhang X, Kang YF, Huang Y, Chen J, Wang G, Zhou Y, Chen Y, Feng QS, Yu H, Li S, Zeng MS, Zeng YX, Xu M, Zheng Q, Chen Y, Zhang X, Xia N
Cell Rep Med (2023) 4 pp. 101296-101296 [ DOI: doi:10.1016/j.xcrm.2023.101296 Pubmed: 37992686 ]
- Ebv ghgl-gp42 in complex with mab 6h2 (Complex)
- 6h2 heavy chain (13 kDa, Protein from Mus musculus)
- Envelope glycoprotein h (34 kDa, Protein from Human gammaherpesvirus 4)
- 6h2 light chain (12 kDa, Protein from Mus musculus)
- 6h2 (Complex from Mus musculus)
- Gh (Complex from Human gammaherpesvirus 4)
Cryo-EM structure of EBV glycoprotein complex gHgL-gp42 bound by a neutralizing antibody 6H2
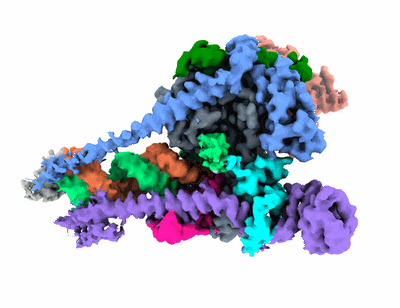
Structure of COVID-19 RNA-dependent RNA polymerase bound to penciclovir.
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7doi
Q-score: 0.599
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Covid-19 rdrp complex bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Pyrophosphate 2- (175 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Water (18 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
Structure of COVID-19 RNA-dependent RNA polymerase (extended conformation) bound to penciclovir
Resolution: 2.73 Å
EM Method: Single-particle
Fitted PDBs: 7dok
Q-score: 0.554
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- [(2r)-4-(2-azanyl-6-oxidanylidene-3h-purin-9-yl)-2-(hydroxymethyl)butyl] dihydrogen phosphate (333 Da, Ligand)
- Water (18 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Covid-19 rdrp complex (exntended conformation) bound to penciclovir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*up*cp*ap*cp*ap*up*ap*gp*c)-3') (7 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna (5'-r(p*gp*cp*up*ap*up*gp*up*gp*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (6 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7dfg
Q-score: 0.598
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Covid-19 rdrp complex bound to favipiravir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Pyrophosphate 2- (175 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Water (18 Da, Ligand)
- Rna (5'-r(p*cp*cp*cp*up*ap*up*ap*ap*cp*up*up*ap*ap*up*cp*u)-3') (4 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 6-fluoro-3-oxo-4-(5-o-phosphono-beta-d-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide (369 Da, Ligand)
- Rna (5'-r(p*ap*gp*ap*up*up*ap*ap*gp*up*up*ap*u)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
Structure of COVID-19 RNA-dependent RNA polymerase bound to ribavirin
Resolution: 2.97 Å
EM Method: Single-particle
Fitted PDBs: 7dfh
Q-score: 0.535
Yin W, Luan X, Li Z, Xie Y, Zhou Z, Liu J, Gao M, Wang X, Zhou F, Wang Q, Wang Q, Shen D, Zhang Y, Tian G, Aisa H, Wei D, Jiang Y, Xiao G, Jiang H, Zhang L, Yu X, Shen J, Zhang S, Xu H
To Be Published
- Covid-19 rdrp complex bound to favipiravir (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rna-directed rna polymeras (107 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Magnesium ion (24 Da, Ligand)
- Pyrophosphate 2- (175 Da, Ligand)
- Water (18 Da, Ligand)
- Rna (5'-r(p*gp*cp*up*ap*up*gp*up*g*(lig))-3') (2 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Ribavirin monophosphate (324 Da, Ligand)
- Rna (5'-r(p*cp*cp*cp*cp*cp*ap*cp*ap*up*ap*gp*c)-3') (3 kDa, RNA from Severe acute respiratory syndrome coronavirus 2)