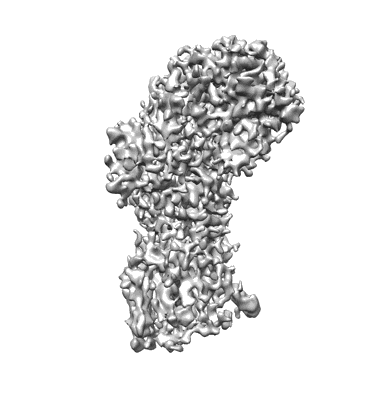
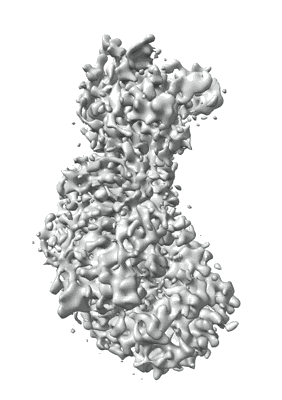
TRIF TIR Filament Cryo-EM Structure
Resolution: 3.3 Å
EM Method: Helical reconstruction
Fitted PDBs: 9dk8
Q-score: 0.47
CryoEM structure of the TIR domain from human TRAM
Resolution: 5.6 Å
EM Method: Helical reconstruction
Fitted PDBs: 9dlg
Q-score: 0.161
Cryo-EM Structure of Full-Length Spike Protein of Omicron XBB.1.5
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 8usz
Q-score: 0.483
Modjarrad K, Che Y, Chen W, Wu H, Cadima CI, Muik A, Maddur MS, Tompkins KR, Martinez LT, Cai H, Ramos M, Mensah S, Cumbia B, Falcao L, McKeen AP, Chang JS, Fennell KF, Huynh KW, McLellan TJ, Sahasrabudhe PV, Chen W, Cerswell M, Garcia MA, Li S, Sharma R, Li W, Dizon KP, Duarte S, Gillett F, Smith R, Illenberger DM, Efferen KS, Vogel AB, Anderson AS, Sahin U, Swanson KA
NPJ Vaccines (2024) 9 pp. 229-229 [ DOI: doi:10.1038/s41541-024-01013-9 Pubmed: 39567521 ]
Cryo-EM structure of CCR6 bound by SQA1 and OXM2
Resolution: 3.02 Å
EM Method: Single-particle
Fitted PDBs: 9d3e
Q-score: 0.499
Wasilko DJ, Gerstenberger BS, Farley KA, Li W, Alley J, Schnute ME, Unwalla RJ, Victorino J, Crouse KK, Ding R, Sahasrabudhe PV, Vincent F, Frisbie RK, Dermenci A, Flick A, Choi C, Chinigo G, Mousseau JJ, Trujillo JI, Nuhant P, Mondal P, Lombardo V, Lamb D, Hogan BJ, Minhas GS, Segala E, Oswald C, Windsor IW, Han S, Rappas M, Cooke RM, Calabrese MF, Berstein G, Thorarensen A, Wu H
Nat Commun (2024) 15 pp. 7574-7574 [ DOI: doi:10.1038/s41467-024-52045-7 Pubmed: 39217154 ]
- Ccr6-bril/fab/nb in complex with sqa1 and oxm2 (120 MDa, Complex from Homo sapiens)
- Human ccr6 (54 kDa, Protein from Homo sapiens)
- Anti-bril fab nanobody (15 kDa, Protein from synthetic construct)
- Anti-bril fab light chain (25 kDa, Protein from Homo sapiens)
- N-{[(2r)-4-(bicyclo[1.1.1]pentan-1-yl)-5-oxomorpholin-2-yl]methyl}-1-[4-(trifluoromethyl)phenyl]cyclopropane-1-carboxamide (408 Da, Ligand)
- Cholesterol (386 Da, Ligand)
- 4-[[3,4-bis(oxidanylidene)-2-[[(1~{r})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{n},~{n}-dimethyl-3-oxidanyl-pyridine-2-carboxamide (426 Da, Ligand)
- Anti-bril fab heavy chain (28 kDa, Protein from Homo sapiens)
Cryo-EM structure of CCR6 bound by SQA1 and OXM1
Wasilko DJ, Gerstenberger BS, Farley KA, Li W, Alley J, Schnute ME, Unwalla RJ, Victorino J, Crouse KK, Ding R, Sahasrabudhe PV, Vincent F, Frisbie RK, Dermenci A, Flick A, Choi C, Chinigo G, Mousseau JJ, Trujillo JI, Nuhant P, Mondal P, Lombardo V, Lamb D, Hogan BJ, Minhas GS, Segala E, Oswald C, Windsor IW, Han S, Rappas M, Cooke RM, Calabrese MF, Berstein G, Thorarensen A, Wu H
Nat Commun (2024) 15 pp. 7574-7574 [ DOI: doi:10.1038/s41467-024-52045-7 Pubmed: 39217154 ]
- Ccr6, soluble cytochrome b562 (54 kDa, Protein from Homo sapiens)
- 1-(4-chlorophenyl)-n-{[(2r)-4-(2,3-dihydro-1h-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide (424 Da, Ligand)
- Anti-bril fab nanobody (15 kDa, Protein from synthetic construct)
- Anti-bril fab light chain (25 kDa, Protein from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- Ccr6-bril/fab/nb in complex with sqa1 and oxm1 (120 MDa, Complex from Homo sapiens)
- 4-[[3,4-bis(oxidanylidene)-2-[[(1~{r})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{n},~{n}-dimethyl-3-oxidanyl-pyridine-2-carboxamide (426 Da, Ligand)
- Anti-bril fab heavy chain (28 kDa, Protein from Homo sapiens)
Caspase and interleukin complex
Devant P, Dong Y, Mintseris J, Ma W, Gygi SP, Wu H, Kagan JC
Nature (2023) 624 pp. 451-459 [ Pubmed: 37993712 DOI: doi:10.1038/s41586-023-06751-9 ]
- Caspase in complex with interleukin (144 kDa, Complex from Homo sapiens)
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens (UltrAuFoil)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8hsb
Q-score: 0.38
Yan Y, Xiao J, Huang F, Xian W, Yu B, Cheng R, Wu H, Lu X, Wang X, Huang W, Li J, Oyejobi GK, Robinson CV, Wu H, Wu D, Liu X, Wang L, Zhu B
Nat Microbiol (2024) 9 pp. 1566-1578 [ DOI: doi:10.1038/s41564-024-01684-z Pubmed: 38649411 ]
- Cdng-e2 binary complex (Complex from Serratia marcescens)
- Type vi secretion protein (18 kDa, Protein from Serratia marcescens)
- Cdng (45 kDa, Protein from Serratia marcescens)
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8yjy
Q-score: 0.426
Yan Y, Xiao J, Huang F, Xian W, Yu B, Cheng R, Wu H, Lu X, Wang X, Huang W, Li J, Oyejobi GK, Robinson CV, Wu H, Wu D, Liu X, Wang L, Zhu B
Nat Microbiol (2024) 9 pp. 1566-1578 [ DOI: doi:10.1038/s41564-024-01684-z Pubmed: 38649411 ]
- Type vi secretion protein (18 kDa, Protein from Serratia marcescens)
- Cdng-e2 binary complex (Complex from Serratia marcescens)
- Smcdng (45 kDa, Protein from Serratia marcescens)
CryoEM structure of nuclear GAPDH under 8h Oxidative Stress
Resolution: 2.17 Å
EM Method: Single-particle
Fitted PDBs: 8g12
Q-score: 0.585
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress
Resolution: 2.3 Å
EM Method: Single-particle
Fitted PDBs: 8g13
Q-score: 0.633
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)