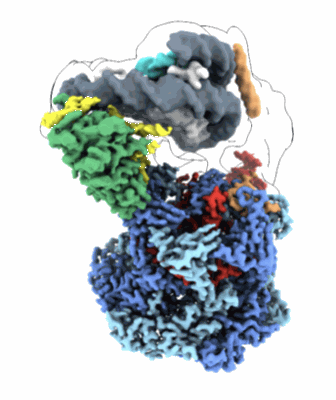
CryoEM structure of nuclear GAPDH under 8h Oxidative Stress
Resolution: 2.17 Å
EM Method: Single-particle
Fitted PDBs: 8g12
Q-score: 0.585
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
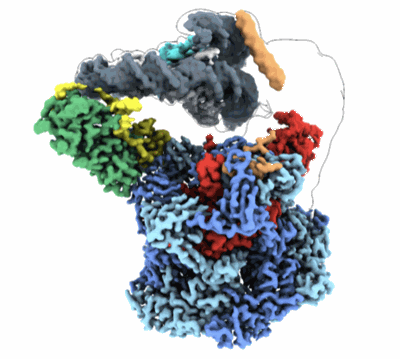
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress
Resolution: 2.3 Å
EM Method: Single-particle
Fitted PDBs: 8g13
Q-score: 0.633
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress, class2
Resolution: 2.3 Å
EM Method: Single-particle
Fitted PDBs: 8g14
Q-score: 0.646
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
CryoEM structure of nuclear GAPDH under 24h Oxidative Stress
Resolution: 2.07 Å
EM Method: Single-particle
Fitted PDBs: 8g15
Q-score: 0.657
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
CryoEM structure of cytoplasmic GAPDH under 24h Oxidative Stress
Resolution: 2.07 Å
EM Method: Single-particle
Fitted PDBs: 8g16
Q-score: 0.653
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
CryoEM structure of wild-type GAPDH
Resolution: 1.98 Å
EM Method: Single-particle
Fitted PDBs: 8g17
Q-score: 0.75
Choi W, Wu H, Yserentant K, Huang B, Cheng Y
PNAS (2023) 120 pp. e2302471120-e2302471120 [ Pubmed: 37487103 DOI: doi:10.1073/pnas.2302471120 ]
- Gapdh (Complex from Homo sapiens)
- Glyceraldehyde-3-phosphate dehydrogenase (35 kDa, Protein from Homo sapiens)
Class1 of the INO80-Hexasome complex
Resolution: 3.04 Å
EM Method: Single-particle
Fitted PDBs: 8ets
Q-score: 0.473
Wu H, Munoz EN, Hsieh LJ, Chio US, Gourdet MA, Narlikar GJ, Cheng Y
Science (2023) 381 pp. 319-324 [ DOI: doi:10.1126/science.adf4197 Pubmed: 37384669 ]
- Actin-related protein 5 (86 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino eighty subunit 2 (3 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 1 (48 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Chromatin-remodeling atpase ino80 (54 kDa, Protein from Saccharomyces cerevisiae S288C)
- Chromatin-remodeling complex subunit ies6 (15 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 2 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino80-hex (Complex from Saccharomyces cerevisiae)
Class2 of the INO80-Hexasome complex
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8etu
Q-score: 0.512
Wu H, Munoz EN, Hsieh LJ, Chio US, Gourdet MA, Narlikar GJ, Cheng Y
Science (2023) 381 pp. 319-324 [ DOI: doi:10.1126/science.adf4197 Pubmed: 37384669 ]
- Actin-related protein 5 (86 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino eighty subunit 2 (3 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 1 (48 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Chromatin-remodeling atpase ino80 (54 kDa, Protein from Saccharomyces cerevisiae S288C)
- Chromatin-remodeling complex subunit ies6 (15 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 2 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino80-hex (Complex from Saccharomyces cerevisiae)
Class3 of INO80-Hexasome complex
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8etw
Q-score: 0.542
Wu H, Munoz EN, Hsieh LJ, Chio US, Gourdet MA, Narlikar GJ, Cheng Y
Science (2023) 381 pp. 319-324 [ DOI: doi:10.1126/science.adf4197 Pubmed: 37384669 ]
- Actin-related protein 5 (86 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino eighty subunit 2 (3 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 1 (48 kDa, Protein from Saccharomyces cerevisiae S288C)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Chromatin-remodeling atpase ino80 (54 kDa, Protein from Saccharomyces cerevisiae S288C)
- Chromatin-remodeling complex subunit ies6 (15 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ruvb-like protein 2 (50 kDa, Protein from Saccharomyces cerevisiae S288C)
- Ino80-hex (Complex from Saccharomyces cerevisiae)
Class1 of the INO80-Hexasome complex
Resolution: 6.68 Å
EM Method: Single-particle
Fitted PDBs: 8ett
Q-score: 0.186
Wu H, Munoz EN, Hsieh LJ, Chio US, Gourdet MA, Narlikar GJ, Cheng Y
Science (2023) 381 pp. 319-324 [ DOI: doi:10.1126/science.adf4197 Pubmed: 37384669 ]
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Dna (110-mer) (70 kDa, DNA from synthetic construct)
- Histone h3.2 (15 kDa, Protein from Xenopus laevis)
- Dna (110-mer) (69 kDa, DNA from synthetic construct)
- Histone h2a type 1 (14 kDa, Protein from Xenopus laevis)
- Class1 hexasome in ino80-hexasome complex (Complex from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)