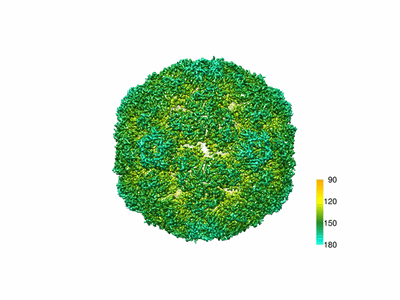
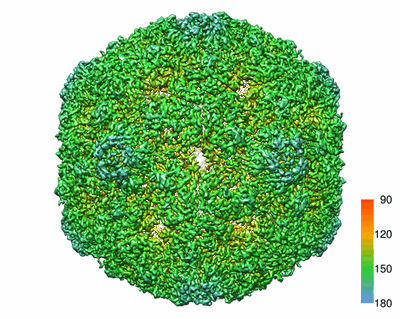
Cryo-EM model of the marine siphophage vB_DshS-R4C portal-adaptor complex
Resolution: 4.7 Å
EM Method: Single-particle
Fitted PDBs: 8gtd
Q-score: 0.238
Cryo-EM structure of pseudorabies virus C-capsid
Resolution: 4.43 Å
EM Method: Single-particle
Fitted PDBs: 7fj1
Q-score: 0.142
Wang G, Zha Z, Huang P, Sun H, Huang Y, He M, Chen T, Lin L, Chen Z, Kong Z, Que Y, Li T, Gu Y, Yu H, Zhang J, Zheng Q, Chen Y, Li S, Xia N
Nat Commun (2022) 13 pp. 1533-1533 [ Pubmed: 35318331 DOI: doi:10.1038/s41467-022-29250-3 ]
- Capsid vertex component 1 (64 kDa, Protein from Suid alphaherpesvirus 1)
- Dna packaging tegument protein ul25 (57 kDa, Protein from Suid alphaherpesvirus 1)
- Vp1/2 (7 kDa, Protein from Suid alphaherpesvirus 1)
- Small capsomere-interacting protein (11 kDa, Protein from Suid alphaherpesvirus 1)
- Triplex capsid protein 1 (40 kDa, Protein from Suid alphaherpesvirus 1)
- Suid alphaherpesvirus 1 (Virus from Suid alphaherpesvirus 1)
- Major capsid protein (146 kDa, Protein from Suid alphaherpesvirus 1)
- Triplex capsid protein 2 (31 kDa, Protein from Suid alphaherpesvirus 1)
Cryo-EM structure of PRV A-capid
Resolution: 4.53 Å
EM Method: Single-particle
Fitted PDBs: 7fj3
Q-score: 0.023
Wang G, Zha Z, Huang P, Sun H, Huang Y, He M, Chen T, Lin L, Chen Z, Kong Z, Que Y, Li T, Gu Y, Yu H, Zhang J, Zheng Q, Chen Y, Li S, Xia N
Nat Commun (2022) 13 pp. 1533-1533 [ Pubmed: 35318331 DOI: doi:10.1038/s41467-022-29250-3 ]
- Small capsomere-interacting protein (11 kDa, Protein from Suid alphaherpesvirus 1)
- Triplex capsid protein 1 (40 kDa, Protein from Suid alphaherpesvirus 1)
- Suid alphaherpesvirus 1 (Virus from Suid alphaherpesvirus 1)
- Major capsid protein (146 kDa, Protein from Suid alphaherpesvirus 1)
- Triplex capsid protein 2 (31 kDa, Protein from Suid alphaherpesvirus 1)
Resolution: 4.19 Å
EM Method: Single-particle
Fitted PDBs: 7dnl
Q-score: 0.339
He M, Chi X, Zha Z, Li Y, Chen J, Huang Y, Huang S, Yu M, Wang Z, Song S, Liu X, Wei S, Li Z, Li T, Wang Y, Yu H, Zhao Q, Zhang J, Zheng Q, Gu Y, Li S, Xia N
J Virol (2021) 95 [ Pubmed: 33472937 DOI: doi:10.1128/JVI.01587-20 ]
- The heavy chain of 2h3 fab fragment (23 kDa, Protein from Mus musculus)
- Fab fragment of a4b4 (Complex from Human papillomavirus type 58)
- Major capsid protein l1 (59 kDa, Protein from Human papillomavirus type 58)
- Human papillomavirus type 58 (Complex)
- The light chain of a4b4 fab fragment (24 kDa, Protein from Mus musculus)
- Human papillomavirus type 58 in complexed with the fab fragment of a4b4 (Complex from Mus musculus)
The atomic structure of varicella-zoster virus A-capsid
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 6lgl
Q-score: 0.114
Wang W, Zheng Q, Pan D, Yu H, Fu W, Liu J, He M, Zhu R, Cai Y, Huang Y, Zha Z, Chen Z, Ye X, Han J, Que Y, Wu T, Zhang J, Li S, Zhu H, Zhou ZH, Cheng T, Xia N
Nat Microbiol (2020) 5 pp. 1542-1552 [ DOI: doi:10.1038/s41564-020-0785-y Pubmed: 32895526 ]
- Triplex capsid protein 1 (54 kDa, Protein from Human herpesvirus 3)
- Human alphaherpesvirus 3 (Virus from Human alphaherpesvirus 3)
- Small capsomere-interacting protein (24 kDa, Protein from Human herpesvirus 3)
- Major capsid protein (155 kDa, Protein from Human herpesvirus 3)
- Triplex capsid protein 2 (34 kDa, Protein from Human herpesvirus 3)
The structure of CVA10 virus procapsid particle
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6acw
Q-score: 0.476
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ Pubmed: 30255146 DOI: doi:10.1126/sciadv.aat7459 ]
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp0 (35 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
The structure of CVA10 virus A-particle from its complex with Fab 2G8
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 6acz
Q-score: 0.466
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ Pubmed: 30255146 DOI: doi:10.1126/sciadv.aat7459 ]
- Vp2 (27 kDa, Protein from Coxsackievirus A10)
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
The structure of CVA10 procapsid from its complex with Fab 2G8
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 6ad1
Q-score: 0.481
Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N
Sci Adv (2018) 4 pp. eaat7459-eaat7459 [ Pubmed: 30255146 DOI: doi:10.1126/sciadv.aat7459 ]
- Vp3 (26 kDa, Protein from Coxsackievirus A10)
- Vp0 (35 kDa, Protein from Coxsackievirus A10)
- Vp1 (33 kDa, Protein from Coxsackievirus A10)
- Coxsackievirus a10 (Virus from Coxsackievirus A10)
The structure of ICAM-5 triggered Enterovirus D68 virus A-particle
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 6aj2
Q-score: 0.412
Zheng Q, Zhu R, Xu L, He M, Yan X, Liu D, Yin Z, Wu Y, Li Y, Yang L, Hou W, Li S, Li Z, Chen Z, Li Z, Yu H, Gu Y, Zhang J, Baker TS, Zhou ZH, Graham BS, Cheng T, Li S, Xia N
Nat Microbiol (2019) 4 pp. 124-133 [ DOI: doi:10.1038/s41564-018-0275-7 Pubmed: 30397341 ]
- Enterovirus d (Virus from Enterovirus D)
- Capsid protein vp1 (32 kDa, Protein from Enterovirus D68)
- Capsid protein vp2 (27 kDa, Protein from Enterovirus D68)
- Capsid protein vp3 (27 kDa, Protein from Enterovirus D68)