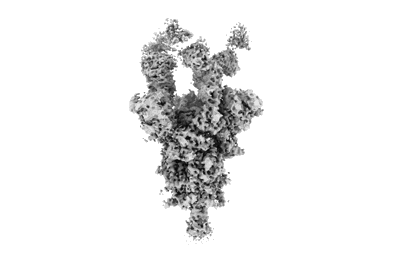
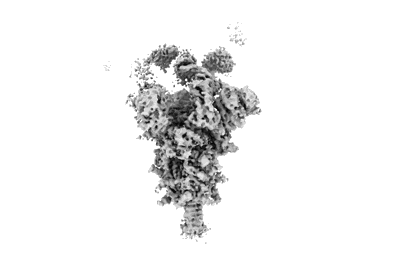
Resolution: 3.15 Å
EM Method: Single-particle
Fitted PDBs: 9dyh
Q-score: 0.476
Owji AP, Dong J, Kittredge A, Wang J, Zhang Y, Yang T
Nat Commun (2024) 15 pp. 10766-10766 [ Pubmed: 39737942 DOI: doi:10.1038/s41467-024-54938-z ]
- Bestrophin-2a (57 kDa, Protein from Homo sapiens)
- Calcium ion (40 Da, Ligand)
- Best2 + glutamate open state (286 kDa, Complex from Homo sapiens)
BEST2 + glutamate closed state
Resolution: 3.03 Å
EM Method: Single-particle
Fitted PDBs: 9dyi
Q-score: 0.555
Owji AP, Dong J, Kittredge A, Wang J, Zhang Y, Yang T
Nat Commun (2024) 15 pp. 10766-10766 [ Pubmed: 39737942 DOI: doi:10.1038/s41467-024-54938-z ]
- Best2 + glutamate closed state (286 kDa, Complex from Homo sapiens)
- Bestrophin-2a (57 kDa, Protein from Homo sapiens)
- Chloride ion (35 Da, Ligand)
- Calcium ion (40 Da, Ligand)
- Glutamic acid (147 Da, Ligand)
Cryo-EM structure of human HCN3 channel with cAMP
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 8io0
Q-score: 0.42
Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7x1y
Q-score: 0.54
Han X, Zhang D, Hong L, Yu D, Wu Z, Yang T, Rust M, Tu Y, Ouyang Q
Nat Commun (2023) 14 pp. 5907- [ DOI: doi:10.1038/s41467-023-41575-1 ]
- Kaic hexamer (Complex from Synechococcus elongatus PCC 7942 = FACHB-805)
- Circadian clock oscillator protein kaic (52 kDa, Protein from Synechococcus elongatus PCC 7942 = FACHB-805)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation
Resolution: 3.53 Å
EM Method: Single-particle
Fitted PDBs: 8heb
Q-score: 0.379
Chu X, Ding X, Yang Y, Lu Y, Li T, Gao Y, Zheng L, Xiao H, Yang T, Cheng H, Huang H, Liu Y, Lou Y, Wu C, Chen Y, Yang H, Ji X, Guo H
Biochem Biophys Res Commun (2023) 660 pp. 43-49 [ DOI: doi:10.1016/j.bbrc.2023.04.002 Pubmed: 37062240 ]
- Rabbit antibody 9h1 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rabbit antibody 9h1 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- Sars-cov-2 wt spike glycoprotein complex with rmab 9h1 fabs (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 9h1 fab (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8hec
Q-score: 0.358
Chu X, Ding X, Yang Y, Lu Y, Li T, Gao Y, Zheng L, Xiao H, Yang T, Cheng H, Huang H, Liu Y, Lou Y, Wu C, Chen Y, Yang H, Ji X, Guo H
Biochem Biophys Res Commun (2023) 660 pp. 43-49 [ DOI: doi:10.1016/j.bbrc.2023.04.002 Pubmed: 37062240 ]
- Rabbit antibody 9h1 light chain (11 kDa, Protein from Oryctolagus cuniculus)
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Rabbit antibody 9h1 heavy chain (12 kDa, Protein from Oryctolagus cuniculus)
- Sars-cov-2 wt spike glycoprotein complex with rmab 9h1 fabs (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 9h1 fab (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 7x1z
Q-score: 0.5
Han X, Zhang D, Hong L, Yu D, Wu Z, Yang T, Rust M, Tu Y, Ouyang Q
Nat Commun (2023) 14 pp. 5907- [ DOI: doi:10.1038/s41467-023-41575-1 ]
- Kaic hexamer (Complex from Synechococcus elongatus PCC 7942 = FACHB-805)
- Magnesium ion (24 Da, Ligand)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Circadian clock oscillator protein kaic (54 kDa, Protein from Synechococcus elongatus)
hBest1 Ca2+-unbound closed state
Resolution: 3.11 Å
EM Method: Single-particle
Fitted PDBs: 8d1m
Q-score: 0.54
Owji AP, Wang J, Kittredge A, Clark Z, Zhang Y, Hendrickson WA, Yang T
Nat Commun (2022) 13 pp. 3836-3836 [ Pubmed: 35789156 DOI: doi:10.1038/s41467-022-31437-7 ]
- Bestrophin-1 (67 kDa, Protein from Homo sapiens)
- Hbest1 ca2+-unbound closed state (338 kDa, Complex from Homo sapiens)
Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7ws4
Q-score: 0.419
Guo H, Gao Y, Li T, Li T, Lu Y, Zheng L, Liu Y, Yang T, Luo F, Song S, Wang W, Yang X, Nguyen HC, Zhang H, Huang A, Jin A, Yang H, Rao Z, Ji X
Cell Rep (2022) 39 pp. 110770-110770 [ DOI: doi:10.1016/j.celrep.2022.110770 Pubmed: 35477022 ]
- 510a5 light chain (11 kDa, Protein from Homo sapiens)
- Omicron spike glycoprotein (Complex from Homo sapiens)
- 510a5 heavy chain (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab (Complex)
- Sars-cov-2 omicron spike glycoprotein complex with 510a5 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development
Resolution: 3.7 Å
EM Method: Single-particle
Fitted PDBs: 7ws5
Q-score: 0.378
Guo H, Gao Y, Li T, Li T, Lu Y, Zheng L, Liu Y, Yang T, Luo F, Song S, Wang W, Yang X, Nguyen HC, Zhang H, Huang A, Jin A, Yang H, Rao Z, Ji X
Cell Rep (2022) 39 pp. 110770-110770 [ DOI: doi:10.1016/j.celrep.2022.110770 Pubmed: 35477022 ]
- 510a5 light chain (11 kDa, Protein from Homo sapiens)
- Omicron spike glycoprotein (Complex from Homo sapiens)
- 510a5 heavy chain (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (133 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab (Complex)
- Sars-cov-2 omicron spike glycoprotein complex with 510a5 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)