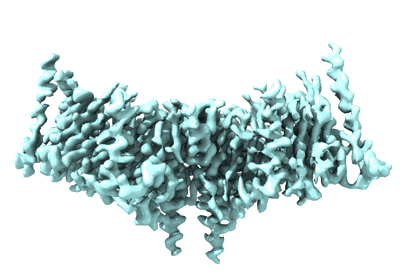
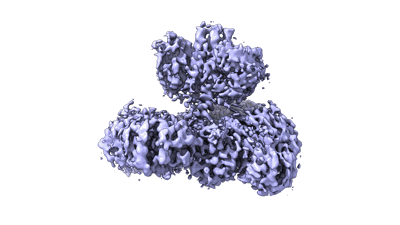
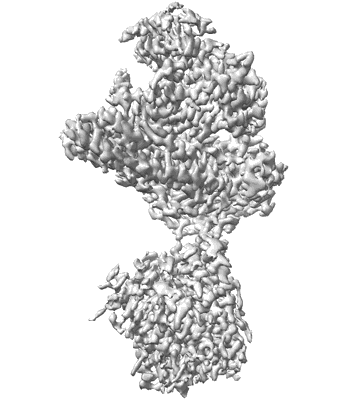
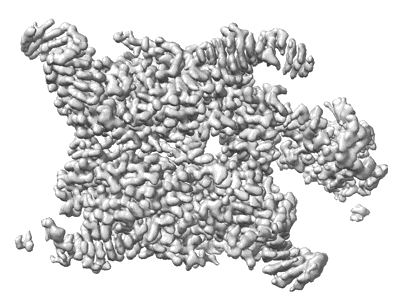
CryoEM structure of human XPR1 in apo state
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 9j4x
Q-score: 0.554
Zhang W, Chen Y, Guan Z, Wang Y, Tang M, Du Z, Zhang J, Cheng M, Zuo J, Liu Y, Wang Q, Liu Y, Zhang D, Yin P, Ma L, Liu Z
Nat Commun (2025) 16 pp. 18-18 [ DOI: doi:10.1038/s41467-024-55471-9 Pubmed: 39747008 ]
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Solute carrier family 53 member 1 (84 kDa, Protein from Homo sapiens)
- Human xpr1 (Complex from Homo sapiens)
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8z9a
Q-score: 0.54
Wang Y, Qiu L, Wang B, Guan Z, Dong Z, Zhang J, Cao S, Yang L, Wang B, Gong Z, Zhang L, Ma W, Liu Z, Zhang D, Wang G, Yin P
Science (2024) 384 pp. 1453-1460 [ Pubmed: 38870272 DOI: doi:10.1126/science.adn6881 ]
- Odorant receptor, apisorco (52 kDa, Protein from Acyrthosiphon pisum)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Or-orco heterocomplex (Complex from Acyrthosiphon pisum)
- Odorant receptor, apisor5 (42 kDa, Protein from Acyrthosiphon pisum)
- [(2e)-3,7-dimethylocta-2,6-dienyl] ethanoate (196 Da, Ligand)
Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8z9z
Q-score: 0.448
Wang Y, Qiu L, Wang B, Guan Z, Dong Z, Zhang J, Cao S, Yang L, Wang B, Gong Z, Zhang L, Ma W, Liu Z, Zhang D, Wang G, Yin P
Science (2024) 384 pp. 1453-1460 [ Pubmed: 38870272 DOI: doi:10.1126/science.adn6881 ]
- Odorant receptor, apisorco (52 kDa, Protein from Acyrthosiphon pisum)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Odorant receptor, apisor5 (42 kDa, Protein from Acyrthosiphon pisum)
- Or-orco heterocomplex (Complex from Acyrthosiphon pisum)
Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8w5k
Q-score: 0.436
Wang Q, Zhuang J, Huang R, Guan Z, Yan L, Hong S, Zhang L, Huang C, Liu Z, Yin P
Cell Discov (2024) 10 pp. 19-19 [ DOI: doi:10.1038/s41421-023-00643-y Pubmed: 38360717 ]
- Mitochondrial import receptor subunit tom5 (5 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom7 (6 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- (2r)-3-{[(s)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate (635 Da, Ligand)
- Mitochondrial import receptor subunit tom40 (42 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom22 (16 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Tom-tim23 complex (Complex from Saccharomyces cerevisiae S288C)
- Mitochondrial import receptor subunit tom6 (6 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex)
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 8w5j
Q-score: 0.281
Wang Q, Zhuang J, Huang R, Guan Z, Yan L, Hong S, Zhang L, Huang C, Liu Z, Yin P
Cell Discov (2024) 10 pp. 19-19 [ DOI: doi:10.1038/s41421-023-00643-y Pubmed: 38360717 ]
- Tom-tim23 complex (Complex from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom5 (5 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom7 (6 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- (2r)-3-{[(s)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate (635 Da, Ligand)
- Mitochondrial import receptor subunit tom40 (42 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom22 (16 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
- Mitochondrial import receptor subunit tom6 (6 kDa, Protein from Saccharomyces cerevisiae (strain ATCC 204508 / S288c))
Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8j5d
Q-score: 0.473
Liu J, Wang X, Guan Z, Wu M, Wang X, Fan R, Zhang F, Yan J, Liu Y, Zhang D, Yin P, Yan J
Plant Cell (2023) 36 pp. 194-212 [ DOI: doi:10.1093/plcell/koad259 Pubmed: 37804098 ]
- Beta-amylase 1, chloroplastic (56 kDa, Protein from Arabidopsis thaliana)
- Malate dehydrogenase, chloroplastic (34 kDa, Protein from Arabidopsis thaliana)
- Bam1-lsf1-mdh complex (Complex from Arabidopsis thaliana)
- Phosphoglucan phosphatase lsf1, chloroplastic (59 kDa, Protein from Arabidopsis thaliana)
Cryo-EM structure of Arabidopsis phytochrome A.
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8iff
Q-score: 0.405
Wang J, Zhou C, Guan Z, Wang Q, Zhao J, Wang L, Zhang L, Zhang D, Deng XW, Ma L, Yin P
Cell Res (2023) 33 pp. 802-805 [ Pubmed: 37402899 DOI: doi:10.1038/s41422-023-00847-7 ]
- 3-[5-[[(3~{r},4~{r})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{h}-pyrrol-2-yl]methyl]-4-methyl-1~{h}-pyrrol-3-yl]propanoic acid (586 Da, Ligand)
- Phytochrome a (Complex from Arabidopsis thaliana)
- Phytochrome a (124 kDa, Protein from Arabidopsis thaliana)
Cryo-EM structure of VTC complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7ytj
Q-score: 0.485
Guan Z, Chen J, Liu R, Chen Y, Xing Q, Du Z, Cheng M, Hu J, Zhang W, Mei W, Wan B, Wang Q, Zhang J, Cheng P, Cai H, Cao J, Zhang D, Yan J, Yin P, Hothorn M, Liu Z
Nat Commun (2023) 14 pp. 718-718 [ DOI: doi:10.1038/s41467-023-36466-4 Pubmed: 36759618 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Vacuolar transporter chaperone 3 (100 kDa, Protein from Saccharomyces cerevisiae)
- Inositol hexakisphosphate (660 Da, Ligand)
- Vtc complex (Complex from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 4 (86 kDa, Protein from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 1 (15 kDa, Protein from Saccharomyces cerevisiae)
Structure of WTAP-VIRMA in the m6A writer complex
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7yg4
Q-score: 0.471
Yan X, Pei K, Guan Z, Liu F, Yan J, Jin X, Wang Q, Hou M, Tang C, Yin P
Cell Res (2022) 32 pp. 1124-1127 [ Pubmed: 36357785 DOI: doi:10.1038/s41422-022-00741-8 ]
- Wtap-virma complex (Complex from Homo sapiens)
- Protein virilizer homolog (123 kDa, Protein from Homo sapiens)
- Pre-mrna-splicing regulator wtap (34 kDa, Protein from Homo sapiens)
Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom
Resolution: 2.51 Å
EM Method: Single-particle
Fitted PDBs: 7xjg
Q-score: 0.515
Wang Y, Guan Z, Wang C, Nie Y, Chen Y, Qian Z, Cui Y, Xu H, Wang Q, Zhao F, Zhang D, Tao P, Sun M, Yin P, Jin S, Wu S, Zou T
Nat Microbiol (2022) 7 pp. 1480-1489 [ Pubmed: 35982312 DOI: doi:10.1038/s41564-022-01197-7 ]
- Retron-ec86 (rt-msdna-rna) (Complex from Escherichia coli)
- Magnesium ion (24 Da, Ligand)
- Rna (81-mer) (25 kDa, RNA from Escherichia coli)
- Dna (105-mer) (32 kDa, DNA from Escherichia coli)
- Rna-directed dna polymerase from retron ec86 (37 kDa, Protein from Escherichia coli)
- Retron st85 family effector protein (35 kDa, Protein from Escherichia coli)
- Rna (14-mer) (4 kDa, RNA from Escherichia coli)