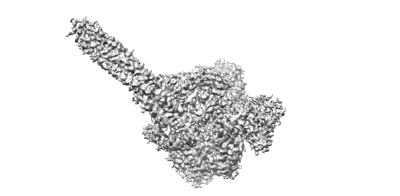
The structure of ASFV DNA polymerase in apo state
Resolution: 3.28 Å
EM Method: Single-particle
Fitted PDBs: 8ywg
Q-score: 0.37
Kuai L, Sun J, Peng Q, Zhao X, Yuan B, Liu S, Bi Y, Shi Y
Nucleic Acids Res (2024) 52 pp. 10717-10729 [ Pubmed: 39189451 DOI: doi:10.1093/nar/gkae739 ]
- Dna polymerase (139 kDa, Protein from African swine fever virus)
- The asfv dna polymerase (Complex from African swine fever virus)
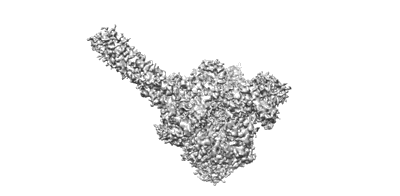
The structure of ASFV DNA polymerase in replicating state
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8ywi
Q-score: 0.508
Kuai L, Sun J, Peng Q, Zhao X, Yuan B, Liu S, Bi Y, Shi Y
Nucleic Acids Res (2024) 52 pp. 10717-10729 [ Pubmed: 39189451 DOI: doi:10.1093/nar/gkae739 ]
- The primer strand (7 kDa, DNA from African swine fever virus)
- Dna polymerase (139 kDa, Protein from African swine fever virus)
- Thymidine-5'-triphosphate (482 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- The template strand (11 kDa, DNA from African swine fever virus)
- The asfv dna polymerase in replicating state (Complex from African swine fever virus)
The structure of ASFV DNA polymerase in editing state
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 8ywm
Q-score: 0.346
Kuai L, Sun J, Peng Q, Zhao X, Yuan B, Liu S, Bi Y, Shi Y
Nucleic Acids Res (2024) 52 pp. 10717-10729 [ Pubmed: 39189451 DOI: doi:10.1093/nar/gkae739 ]
- Dna polymerase (139 kDa, Protein from African swine fever virus)
- The template strand (8 kDa, DNA from African swine fever virus)
- The asfv dna polymerase in editing state (Complex from African swine fever virus)
- The primer strand (6 kDa, DNA from African swine fever virus)
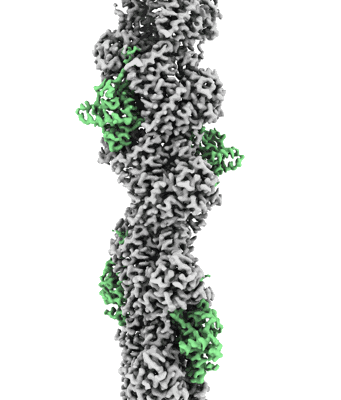
F-actin decorated by SipA497-669
Resolution: 2.7 Å
EM Method: Helical reconstruction
Fitted PDBs: 8c4c
Q-score: 0.648
Yuan B, Scholz J, Wald J, Thuenauer R, Hennell James R, Ellenberg I, Windhorst S, Faix J, Marlovits TC
Sci Adv (2023) 9 pp. eadj5777-eadj5777 [ DOI: doi:10.1126/sciadv.adj5777 Pubmed: 38064550 ]
- Phosphate ion (94 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Cell invasion protein sipa (19 kDa, Protein from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- F-actin (Complex from Gallus gallus)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Actin binding domain sipa497-669 (Complex from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2)
- F-actin polymerized by sipa497-669 under low-salt conditions (Complex)
- Actin, alpha skeletal muscle (42 kDa, Protein from Gallus gallus)
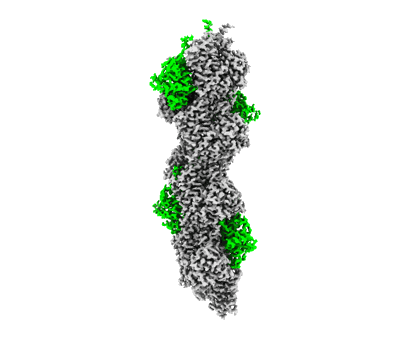
F-actin decorated by SipA426-685
Resolution: 2.6 Å
EM Method: Helical reconstruction
Fitted PDBs: 8c4e
Q-score: 0.676
Yuan B, Scholz J, Wald J, Thuenauer R, Hennell James R, Ellenberg I, Windhorst S, Faix J, Marlovits TC
Sci Adv (2023) 9 pp. eadj5777-eadj5777 [ DOI: doi:10.1126/sciadv.adj5777 Pubmed: 38064550 ]
- Phosphate ion (94 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- F-actin (Complex from Gallus gallus)
- Adenosine-5'-diphosphate (427 Da, Ligand)
- Actin binding domain sipa-c (Complex from Salmonella)
- Cell invasion protein sipa (28 kDa, Protein from Salmonella)
- F-actin polymerized by sipa497-669 under low-salt conditions (Complex)
- Actin, alpha skeletal muscle (42 kDa, Protein from Gallus gallus)
Cryo-EM structure of PcrV/Fab(30-B8)
Resolution: 4.2 Å
EM Method: Single-particle
Fitted PDBs: 8cr9
Q-score: 0.328
Simonis A, Kreer C, Albus A, Rox K, Yuan B, Holzmann D, Wilms JA, Zuber S, Kottege L, Winter S, Meyer M, Schmitt K, Gruell H, Theobald SJ, Hellmann AM, Meyer C, Ercanoglu MS, Cramer N, Munder A, Hallek M, Fatkenheuer G, Koch M, Seifert H, Rietschel E, Marlovits TC, van Koningsbruggen-Rietschel S, Klein F, Rybniker J
Cell (2023) 186 pp. 5098-5113.e19 [ Pubmed: 37918395 DOI: doi:10.1016/j.cell.2023.10.002 ]
- Maltose/maltodextrin-binding periplasmic protein,type iii secretion protein pcrv (75 kDa, Protein from Pseudomonas aeruginosa)
- Heavy chain (23 kDa, Protein from Homo sapiens)
- Pcrv-fab(30-b8) (Complex from Homo sapiens)
- Light chain (23 kDa, Protein from Homo sapiens)
Cryo-EM structure of PcrV/Fab(11-E5)
Resolution: 4.6 Å
EM Method: Single-particle
Fitted PDBs: 8crb
Q-score: 0.308
Simonis A, Kreer C, Albus A, Rox K, Yuan B, Holzmann D, Wilms JA, Zuber S, Kottege L, Winter S, Meyer M, Schmitt K, Gruell H, Theobald SJ, Hellmann AM, Meyer C, Ercanoglu MS, Cramer N, Munder A, Hallek M, Fatkenheuer G, Koch M, Seifert H, Rietschel E, Marlovits TC, van Koningsbruggen-Rietschel S, Klein F, Rybniker J
Cell (2023) 186 pp. 5098-5113.e19 [ Pubmed: 37918395 DOI: doi:10.1016/j.cell.2023.10.002 ]
- Maltose/maltodextrin-binding periplasmic protein,type iii secretion protein pcrv (75 kDa, Protein from Pseudomonas aeruginosa)
- Light chain (22 kDa, Protein from Homo sapiens)
- Pcrv-fab(11-e5) (Complex from Homo sapiens)
- Heavy chain (24 kDa, Protein from Homo sapiens)
The structure of EBOV L-VP35-RNA complex
Resolution: 2.95 Å
EM Method: Single-particle
Fitted PDBs: 8jsl
Q-score: 0.547
Peng Q, Yuan B, Cheng J, Wang M, Gao S, Bai S, Zhao X, Qi J, Gao GF, Shi Y
Nature (2023) 622 pp. 603-610 [ DOI: doi:10.1038/s41586-023-06608-1 Pubmed: 37699521 ]
- The leader sequence of ebov (5 kDa, RNA from Ebola virus)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase l (252 kDa, Protein from Ebola virus)
- The structure of ebov l-vp35-rna complex (Complex from Ebola virus)
- Polymerase cofactor vp35 (37 kDa, Protein from Ebola virus)
The structure of EBOV L-VP35-RNA complex (conformation 1)
Resolution: 3.3 Å
EM Method: Single-particle
Fitted PDBs: 8jsm
Q-score: 0.495
Peng Q, Yuan B, Cheng J, Wang M, Gao S, Bai S, Zhao X, Qi J, Gao GF, Shi Y
Nature (2023) 622 pp. 603-610 [ DOI: doi:10.1038/s41586-023-06608-1 Pubmed: 37699521 ]
- The leader sequence of ebov genome. (5 kDa, RNA from Ebola virus)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase l (252 kDa, Protein from Ebola virus)
- The structure of ebov l-vp35-rna complex (conformation 1) (Complex from Ebola virus)
- Polymerase cofactor vp35 (37 kDa, Protein from Ebola virus)
The structure of EBOV L-VP35-RNA complex (conformation 2)
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 8jsn
Q-score: 0.475
Peng Q, Yuan B, Cheng J, Wang M, Gao S, Bai S, Zhao X, Qi J, Gao GF, Shi Y
Nature (2023) 622 pp. 603-610 [ DOI: doi:10.1038/s41586-023-06608-1 Pubmed: 37699521 ]
- The structure of ebov l-vp35-rna complex (conformation 2) (Complex from Ebola virus)
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase l (252 kDa, Protein from Ebola virus)
- Polymerase cofactor vp35 (37 kDa, Protein from Ebola virus)
- The leader sequence of ebov genome (5 kDa, RNA from Ebola virus)