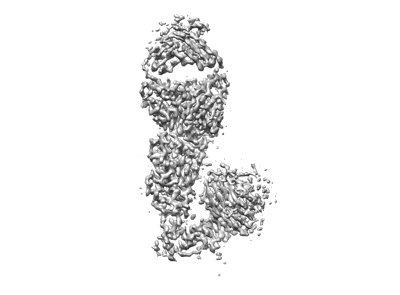
SARS-CoV-2 spike trimer in complex with Fab NE12, local refinement map
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7u9o
Q-score: 0.518
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Ne12 fab heavy chain (24 kDa, Protein from Homo sapiens)
- Ne12 fab light chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike receptor-binding domain in complex with fab ne12 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
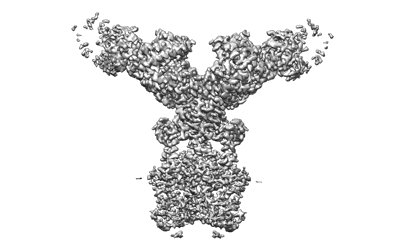
SARS-CoV-2 spike trimer in complex with Fab NA8, local refinement map
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7u9p
Q-score: 0.528
Chen Z, Zhang P, Matsuoka Y, Tsybovsky Y, West K, Santos C, Boyd LF, Nguyen H, Pomerenke A, Stephens T, Olia AS, Zhang B, De Giorgi V, Holbrook MR, Gross R, Postnikova E, Garza NL, Johnson RF, Margulies DH, Kwong PD, Alter HJ, Buchholz UJ, Lusso P, Farci P
Cell Rep (2022) 41 pp. 111528-111528 [ Pubmed: 36302375 DOI: doi:10.1016/j.celrep.2022.111528 ]
- Sars-cov-2 spike receptor-binding domain in complex with fab na8 (110 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Na8 fab light chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Na8 fab heavy chain (24 kDa, Protein from Homo sapiens)
ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7v61
Q-score: 0.366
Chen Y, Zhang YN, Yan R, Wang G, Zhang Y, Zhang ZR, Li Y, Ou J, Chu W, Liang Z, Wang Y, Chen YL, Chen G, Wang Q, Zhou Q, Zhang B, Wang C
Signal Transduct Target Ther (2021) 6 pp. 315-315 [ Pubmed: 34433803 DOI: doi:10.1038/s41392-021-00740-y ]
- 3e8 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Ace2-b0at1 complex (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Zinc ion (65 Da, Ligand)
- Sodium-dependent neutral amino acid transporter b(0)at1 (73 kDa, Protein from Homo sapiens)
- 3e8 (49 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Angiotensin-converting enzyme 2 (93 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
Structure of freshly purified SARS-CoV-2 S2P spike at pH 7.4
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7mtc
Q-score: 0.6
Olia AS, Tsybovsky Y, Chen SJ, Liu C, Nazzari AF, Ou L, Wang L, Kong WP, Leung K, Liu T, Stephens T, Teng IT, Wang S, Yang ES, Zhang B, Zhang Y, Zhou T, Mascola JR, Kwong PD
J Biol Chem (2021) 297 pp. 101127-101127 [ Pubmed: 34461095 DOI: doi:10.1016/j.jbc.2021.101127 ]
Structure of aged SARS-CoV-2 S2P spike at pH 7.4
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7mtd
Q-score: 0.443
Olia AS, Tsybovsky Y, Chen SJ, Liu C, Nazzari AF, Ou L, Wang L, Kong WP, Leung K, Liu T, Stephens T, Teng IT, Wang S, Yang ES, Zhang B, Zhang Y, Zhou T, Mascola JR, Kwong PD
J Biol Chem (2021) 297 pp. 101127-101127 [ Pubmed: 34461095 DOI: doi:10.1016/j.jbc.2021.101127 ]
Structure of SARS-CoV-2 S2P spike at pH 7.4 refolded by low-pH treatment
Resolution: 3.2 Å
EM Method: Single-particle
Fitted PDBs: 7mte
Q-score: 0.523
Olia AS, Tsybovsky Y, Chen SJ, Liu C, Nazzari AF, Ou L, Wang L, Kong WP, Leung K, Liu T, Stephens T, Teng IT, Wang S, Yang ES, Zhang B, Zhang Y, Zhou T, Mascola JR, Kwong PD
J Biol Chem (2021) 297 pp. 101127-101127 [ Pubmed: 34461095 DOI: doi:10.1016/j.jbc.2021.101127 ]
Structure of SARS-CoV-2 spike at pH 4.5
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 7jwy
Q-score: 0.612
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike at ph 4.5 (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
Structure of SARS-CoV-2 spike at pH 4.0
Resolution: 2.4 Å
EM Method: Single-particle
Fitted PDBs: 6xlu
Q-score: 0.63
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Water (18 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Consensus structure of SARS-CoV-2 spike at pH 5.5
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6xm0
Q-score: 0.583
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 6xm3
Q-score: 0.562
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD
Cell Host Microbe (2020) 28 pp. 867-879.e5 [ DOI: doi:10.1016/j.chom.2020.11.004 Pubmed: 33271067 ]
- Spike glycoprotein (140 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike (423 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)