Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: 10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
Image sets:
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: 10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
Image sets:
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: 10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
Image sets:
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: 10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
Image sets:
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: 10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
Image sets:
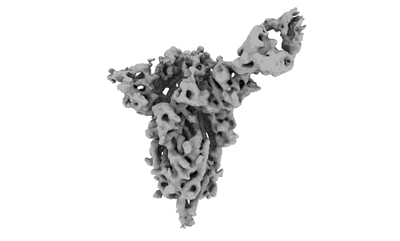
Cryo-EM structure of NTD-directed neutralizing antibody LP5 Fab in complex with SARS-CoV-2 S2P spike
Resolution: 4.46 Å
EM Method: Single-particle
Fitted PDBs: 7mxp
Q-score: 0.192
Madan B, Reddem ER, Wang P, Casner RG, Nair MS, Huang Y, Fahad AS, de Souza MO, Banach BB, Lopez Acevedo SN, Pan X, Nimrania R, Teng IT, Bahna F, Zhou T, Zhang B, Yin MT, Ho DD, Kwong PD, Shapiro L, DeKosky BJ
Aiche J (2021) 67 pp. e17440-e17440 [ DOI: doi:10.1002/aic.17440 Pubmed: 34898670 ]
- Spike glycoprotein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Lp5 fab light chain (11 kDa, Protein from Homo sapiens)
- Lp5 heavy chain, lp5 light chain (Complex from Homo sapiens)
- Prefusion sars-cov-2 spike glycoprotein in complex with lp5 fab (Complex)
- Lp5 fab heavy chain (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ Pubmed: 33789084 DOI: doi:10.1016/j.chom.2021.03.005 ]
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7l56
Q-score: 0.475
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab 2-43 variable domain heavy chain (14 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Fab 2-43 variable domain light chain (11 kDa, Protein from Homo sapiens)
- Cryo-em structure of the sars-cov-2 spike glycoprotein bound to fab 2-43 (Complex from Homo sapiens)
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15
Resolution: 5.87 Å
EM Method: Single-particle
Fitted PDBs: 7l57
Q-score: 0.311
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- Fab 2-15 variable domain light chain (11 kDa, Protein from Homo sapiens)
- Fab 2-15 variable domain heavy chain (14 kDa, Protein from Homo sapiens)
- Cryo-em structure of the sars-cov-2 spike glycoprotein bound to fab 2-15 (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4
Resolution: 5.07 Å
EM Method: Single-particle
Fitted PDBs: 7l58
Q-score: 0.331
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- Fab h4 variable domain light chain (12 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 trimeric spike glycoprotein bound to one copy of fab h4 (Complex from Homo sapiens)
- Fab h4 variable domain heavy chain (13 kDa, Protein from Homo sapiens)