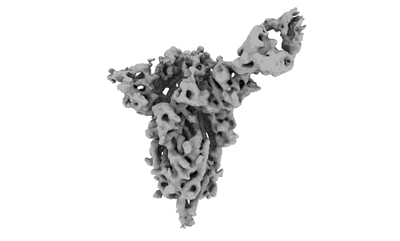
5-HT1A-Gi in complex with compound (R)-IHCH-7179
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8jt6
Q-score: 0.542
Chen Z, Yu J, Wang H, Xu P, Fan L, Sun F, Huang S, Zhang P, Huang H, Gu S, Zhang B, Zhou Y, Wan X, Pei G, Xu HE, Cheng J, Wang S
Cell (2024) 187 pp. 2194-2208.e22 [ Pubmed: 38552625 DOI: doi:10.1016/j.cell.2024.02.034 ]
- Palmitic acid (256 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (37 kDa, Protein from Homo sapiens)
- Soluble cytochrome b562,5-hydroxytryptamine receptor 1a (60 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- [(2r)-1-[oxidanyl-[(2r,3r,5s,6r)-2,3,5,6-tetrakis(oxidanyl)-4-phosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-3-tetradecanoyloxy-propan-2-yl] (5e,8e)-hexadeca-5,8,11,14-tetraenoate (854 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Cholesterol (386 Da, Ligand)
- 1-(4-fluorophenyl)-4-[(7r)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one (377 Da, Ligand)
- Scfv16 (28 kDa, Protein from Mus musculus)
- 5-ht1a-gi in complex with compound (r)-ihch-7179 (Complex from Homo sapiens)
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8ipq
Q-score: 0.332
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8ipr
Q-score: 0.476
Zhu C, Shi Y, Yu J, Zhao W, Li L, Liang J, Yang X, Zhang B, Zhao Y, Gao Y, Chen X, Yang X, Zhang L, Guddat LW, Liu L, Yang H, Rao Z, Li J
Protein Cell (2023) 14 pp. 919-923 [ DOI: doi:10.1093/procel/pwad022 Pubmed: 37144855 ]
- Component linked with the assembly of cytochrome' abc transporter atp-binding protein cydc (51 kDa, Protein from Mycolicibacterium smegmatis)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Transmembrane atp-binding protein abc transporter cydd (53 kDa, Protein from Mycolicibacterium smegmatis)
- Cyddc (Complex from Mycolicibacterium smegmatis)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8ips
Q-score: 0.472
Zhu C, Shi Y, Yu J, Zhao W, Li L, Liang J, Yang X, Zhang B, Zhao Y, Gao Y, Chen X, Yang X, Zhang L, Guddat LW, Liu L, Yang H, Rao Z, Li J
Protein Cell (2023) 14 pp. 919-923 [ DOI: doi:10.1093/procel/pwad022 Pubmed: 37144855 ]
- Abc transporter, cyddc cysteine exporter (cyddc-e) family, permease/atp-binding protein cydc (64 kDa, Protein from Escherichia coli (strain B / BL21-DE3))
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Abc transporter, cyddc cysteine exporter (cyddc-e) family, permease/atp-binding protein cydd (65 kDa, Protein from Escherichia coli (strain B / BL21-DE3))
- Cyddc (Complex from Escherichia coli)
Cryo-EM structure of heme transporter CydDC from Escherichia coli in the occluded ATP bound state
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8ipt
Q-score: 0.456
Zhu C, Shi Y, Yu J, Zhao W, Li L, Liang J, Yang X, Zhang B, Zhao Y, Gao Y, Chen X, Yang X, Zhang L, Guddat LW, Liu L, Yang H, Rao Z, Li J
Protein Cell (2023) 14 pp. 919-923 [ DOI: doi:10.1093/procel/pwad022 Pubmed: 37144855 ]
- Abc transporter, cyddc cysteine exporter (cyddc-e) family, permease/atp-binding protein cydc (62 kDa, Protein from Escherichia coli)
- Abc transporter, cyddc cysteine exporter (cyddc-e) family, permease/atp-binding protein cydd (65 kDa, Protein from Escherichia coli)
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Cyddc (Complex from Escherichia coli)
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819-833.e7 [ DOI: doi:10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
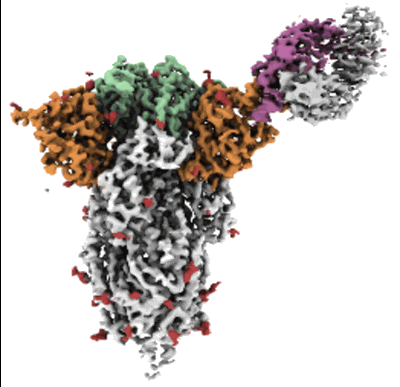
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7l56
Q-score: 0.475
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab 2-43 variable domain heavy chain (14 kDa, Protein from Homo sapiens)
- Cryo-em structure of the sars-cov-2 spike glycoprotein bound to fab 2-43 (Complex from Homo sapiens)
- Fab 2-43 variable domain light chain (11 kDa, Protein from Homo sapiens)
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15
Resolution: 5.87 Å
EM Method: Single-particle
Fitted PDBs: 7l57
Q-score: 0.311
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Fab 2-15 variable domain light chain (11 kDa, Protein from Homo sapiens)
- Fab 2-15 variable domain heavy chain (14 kDa, Protein from Homo sapiens)
- Cryo-em structure of the sars-cov-2 spike glycoprotein bound to fab 2-15 (Complex from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4
Resolution: 5.07 Å
EM Method: Single-particle
Fitted PDBs: 7l58
Q-score: 0.331
Rapp M, Guo Y, Reddem ER, Yu J, Liu L, Wang P, Cerutti G, Katsamba P, Bimela JS, Bahna FA, Mannepalli SM, Zhang B, Kwong PD, Huang Y, Ho DD, Shapiro L, Sheng Z
Cell Rep (2021) 35 pp. 108950-108950 [ Pubmed: 33794145 DOI: doi:10.1016/j.celrep.2021.108950 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 trimeric spike glycoprotein bound to one copy of fab h4 (Complex from Homo sapiens)
- Fab h4 variable domain heavy chain (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Fab h4 variable domain light chain (12 kDa, Protein from Homo sapiens)
Resolution: 3.55 Å
EM Method: Single-particle
Fitted PDBs: 7l2d
Q-score: 0.425
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L
Cell Host Microbe (2021) 29 pp. 819- [ DOI: doi:10.1016/j.chom.2021.03.005 Pubmed: 33789084 ]
- 1-87 light chain (22 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 1-87 heavy chain (25 kDa, Protein from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Prefusion sars-cov-2 spike glycoprotein in complex with 1-87 fab (Complex from Homo sapiens)