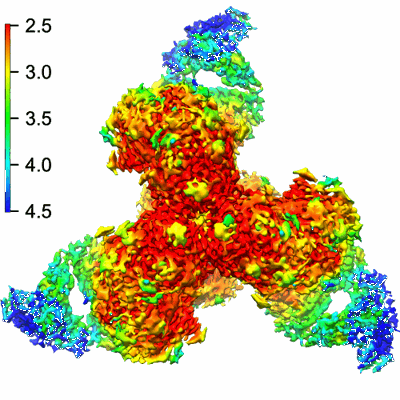
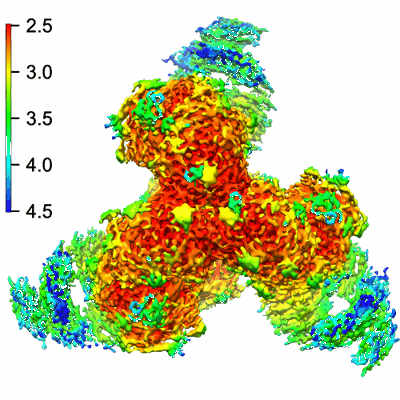
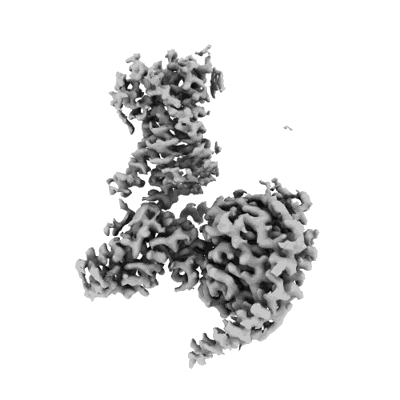
Structure of VRC44.01 Fab in complex with 3BNC117-purified C1080.c3 RnS SOSIP.664 HIV-1 Env trimer
Resolution: 2.83 Å
EM Method: Single-particle
Fitted PDBs: 9bio
Q-score: 0.547
Cale EM, Shen CH, Olia AS, Radakovich NA, Rawi R, Yang Y, Ambrozak DR, Bennici AK, Chuang GY, Crooks ED, Driscoll JI, Lin BC, Louder MK, Madden PJ, Messina MA, Osawa K, Stewart-Jones GBE, Verardi R, Vrakas Z, Xie D, Zhang B, Binley JM, Connors M, Koup RA, Pierson TC, Doria-Rose NA, Kwong PD, Mascola JR, Gorman J
Cell Rep (2024) 43 pp. 115010-115010 [ Pubmed: 39675002 DOI: doi:10.1016/j.celrep.2024.115010 ]
- 3bnc117 light chain (23 kDa, Protein from Homo sapiens)
- Phosphate ion (94 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- 3bnc117 heavy chain (24 kDa, Protein from Homo sapiens)
- Vrc44.01 heavy chain (13 kDa, Protein from Homo sapiens)
- Envelope glycoprotein gp160 (54 kDa, Protein from Human immunodeficiency virus 1)
- Vrc44 and 3bnc117 fabs with repaired c1080 env trimer (Complex from Human immunodeficiency virus 1)
- Vrc44.01 light chain (11 kDa, Protein from Homo sapiens)
- Envelope glycoprotein gp41 (19 kDa, Protein from Human immunodeficiency virus 1)
Cryo-EM structure of BG505 Env mutant A517E in complex with antibody ACS202 Fab
Reveiz M, Xu K, Lee M, Wang S, Olia AS, Harris DR, Liu K, Liu T, Schaub AJ, Stephens T, Wang Y, Zhang B, Huang R, Tsybovsky Y, Kwong PD, Rawi R
Front Immunol (2024) 15 pp. 1484029-1484029 [ DOI: doi:10.3389/fimmu.2024.1484029 Pubmed: 39611147 ]
- Hiv-1 env trimer complexed with antibody fab (Complex from Human immunodeficiency virus 1)
- Envelope glycoprotein gp120 (53 kDa, Protein from Human immunodeficiency virus 1)
- Antibody acs202 fab light chain (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Antibody acs202 fab heavy chain (25 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Antibody fab (Complex from Homo sapiens)
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8euv
Q-score: 0.586
Banach BB, Pletnev S, Olia AS, Xu K, Zhang B, Rawi R, Bylund T, Doria-Rose NA, Nguyen TD, Fahad AS, Lee M, Lin BC, Liu T, Louder MK, Madan B, McKee K, O'Dell S, Sastry M, Schon A, Bui N, Shen CH, Wolfe JR, Chuang GY, Mascola JR, Kwong PD, DeKosky BJ
Nat Commun (2023) 14 pp. 7593-7593 [ Pubmed: 37989731 DOI: doi:10.1038/s41467-023-42098-5 ]
- Vrc34.01-combo1 fab variable heavy chain (23 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Bg505 ds-sosip vrc34.01-combo1 fab complex (Complex from Human immunodeficiency virus 1)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Vrc34.01-combo1 fab variable light chain (23 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Envelope glycoprotein gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8euw
Q-score: 0.553
Banach BB, Pletnev S, Olia AS, Xu K, Zhang B, Rawi R, Bylund T, Doria-Rose NA, Nguyen TD, Fahad AS, Lee M, Lin BC, Liu T, Louder MK, Madan B, McKee K, O'Dell S, Sastry M, Schon A, Bui N, Shen CH, Wolfe JR, Chuang GY, Mascola JR, Kwong PD, DeKosky BJ
Nat Commun (2023) 14 pp. 7593-7593 [ Pubmed: 37989731 DOI: doi:10.1038/s41467-023-42098-5 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Vrc34.01-mm28 fab variable heavy chain (23 kDa, Protein from Homo sapiens)
- Bg505 ds-sosip vrc34.01-mm28 fab complex (Complex from Human immunodeficiency virus 1)
- Envelope glycoprotein gp41 (17 kDa, Protein from Human immunodeficiency virus 1)
- Vrc34.01-mm28 fab variable light chain (23 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Envelope glycoprotein gp120 (54 kDa, Protein from Human immunodeficiency virus 1)
Cryo-EM structure of Cas12g-sgRNA binary complex
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8i3q
Q-score: 0.461
Liu M, Li Z, Chen J, Lin J, Lu Q, Ye Y, Zhang H, Zhang B, Ouyang S
PLoS Genet (2023) 19 pp. e1010930-e1010930 [ Pubmed: 37729124 DOI: doi:10.1371/journal.pgen.1010930 ]
- Rna (Complex)
- Zinc ion (65 Da, Ligand)
- Rna (139-mer) (44 kDa, RNA from Escherichia coli)
- Cas12g (Complex)
- Cas12g-sgrna binary complex (Complex from Escherichia coli)
- Cas12g (89 kDa, Protein from Escherichia coli)
Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7wu4
Q-score: 0.428
Qu X, Qiu N, Wang M, Zhang B, Du J, Zhong Z, Xu W, Chu X, Ma L, Yi C, Han S, Shui W, Zhao Q, Wu B
Nature (2022) 604 pp. 779-785 [ Pubmed: 35418679 DOI: doi:10.1038/s41586-022-04580-w ]
- [(2~{r})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate (496 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Homo sapiens)
- Adhesion g-protein coupled receptor f1 (74 kDa, Protein from Homo sapiens)
- The adhesion gpcr adgrf1 in complex with minigi (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (27 kDa, Protein from Homo sapiens)
- Cholesterol (386 Da, Ligand)
Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7wu5
Q-score: 0.492
Qu X, Qiu N, Wang M, Zhang B, Du J, Zhong Z, Xu W, Chu X, Ma L, Yi C, Han S, Shui W, Zhao Q, Wu B
Nature (2022) 604 pp. 779-785 [ Pubmed: 35418679 DOI: doi:10.1038/s41586-022-04580-w ]
- Adhesion g-protein coupled receptor f1 (74 kDa, Protein from Homo sapiens)
- [(2~{r})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate (496 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- The adhesion gpcr adgrf1(h565a/t567a) in complex with minigi (Complex from Homo sapiens)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (27 kDa, Protein from Homo sapiens)
- Cholesterol (386 Da, Ligand)
S protein of SARS-CoV-2 in complex with 2G1
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7x08
Q-score: 0.458
Ma H, Guo Y, Tang H, Tseng CK, Wang L, Zong H, Wang Z, He Y, Chang Y, Wang S, Huang H, Ke Y, Yuan Y, Wu M, Zhang Y, Drelich A, Kempaiah KR, Peng BH, Wang A, Yang K, Yin H, Liu J, Yue Y, Xu W, Zhu S, Ji T, Zhang X, Wang Z, Li G, Liu G, Song J, Mu L, Xiang Z, Song Z, Chen H, Bian Y, Zhang B, Chen H, Zhang J, Liao Y, Zhang L, Yang L, Chen Y, Gilly J, Xiao X, Han L, Jiang H, Xie Y, Zhou Q, Zhu J
Cell Discov (2022) 8 pp. 16-16 [ DOI: doi:10.1038/s41421-022-00381-7 Pubmed: 35169121 ]
- S protein of sars-cov-2 in complex with 2g1 (Complex)
- Light chain of 2g1 (22 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Heavy chain of 2g1 (49 kDa, Protein from Homo sapiens)
- 2g1 (Complex from Homo sapiens)
- Spike glycoprotein (142 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Linoleic acid (280 Da, Ligand)
- S protein of sars-cov-2 (Complex from Severe acute respiratory syndrome coronavirus)
Resolution: 3.9 Å
EM Method: Single-particle
Fitted PDBs: 6xh7
Q-score: 0.344
Shi W, Zhang B, Jiang Y, Liu C, Zhou W, Chen M, Yang Y, Hu Y, Liu B
iScience (2021) 24 pp. 102449-102449 [ DOI: doi:10.1016/j.isci.2021.102449 Pubmed: 34113812 ]
- Rna polymerase sigma factor rpod (72 kDa, Protein from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Template strand dna (54-mer) (16 kDa, DNA from Escherichia coli)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli)
- Cuer-tac without rna (530 kDa, Complex from Escherichia coli)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli)
- Copper (ii) ion (63 Da, Ligand)
- Nontemplate strand dna (54-mer) (16 kDa, DNA from Escherichia coli)
- Hth-type transcriptional regulator cuer (16 kDa, Protein from Escherichia coli)
CueR-transcription activation complex with RNA transcript
Resolution: 4.1 Å
EM Method: Single-particle
Fitted PDBs: 6xh8
Q-score: 0.318
Shi W, Zhang B, Jiang Y, Liu C, Zhou W, Chen M, Yang Y, Hu Y, Liu B
iScience (2021) 24 pp. 102449-102449 [ DOI: doi:10.1016/j.isci.2021.102449 Pubmed: 34113812 ]
- Rna polymerase sigma factor rpod (72 kDa, Protein from Escherichia coli)
- Zinc ion (65 Da, Ligand)
- Template strand dna (54-mer) (16 kDa, DNA from Escherichia coli)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli)
- Cuer-tac with rna (540 kDa, Complex from Escherichia coli)
- Nascent rna (1 kDa, RNA from Escherichia coli)
- Copper (ii) ion (63 Da, Ligand)
- Nontemplate strand dna (54-mer) (16 kDa, DNA from Escherichia coli)
- Hth-type transcriptional regulator cuer (16 kDa, Protein from Escherichia coli)