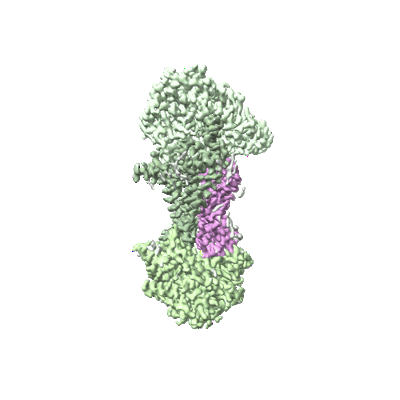
Cryo-EM structure of the ATP-bound DppABCD complex
Resolution: 2.86 Å
EM Method: Single-particle
Fitted PDBs: 8wdb
Q-score: 0.605
To Be Published
- Probable periplasmic dipeptide-binding lipoprotein dppa (56 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Probable dipeptide-transport integral membrane protein abc transporter dppb (32 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Probable dipeptide-transport atp-binding protein abc transporter dppd (59 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Probable dipeptide-transport integral membrane protein abc transporter dppc (30 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Dppabcd (Complex from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the catalytic intermediate state
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 8j5t
Q-score: 0.572
Yang X, Hu T, Liang J, Xiong Z, Lin Z, Zhao Y, Zhou X, Gao Y, Sun S, Yang X, Guddat LW, Yang H, Rao Z, Zhang B
Nat Struct Mol Biol (2024) 31 pp. 1072-1082 [ DOI: doi:10.1038/s41594-024-01256-z Pubmed: 38548954 ]
- Putative peptide transport permease protein rv1283c (35 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Uncharacterized protein rv1280c (64 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Putative peptide transport permease protein rv1282c (31 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Uncharacterized abc transporter atp-binding protein rv1281c (65 kDa, Protein from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Cryo-em structure of mycobacterium tuberculosis oppabcd in the catalytic intermediate state (Complex from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- Adenosine-5'-triphosphate (507 Da, Ligand)
- Magnesium ion (24 Da, Ligand)
- Iron/sulfur cluster (351 Da, Ligand)
Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP
Resolution: 2.88 Å
EM Method: Single-particle
Fitted PDBs: 7wiv
Q-score: 0.577
Sun S, Gao Y, Yang X, Yang X, Hu T, Liang J, Xiong Z, Ran Y, Ren P, Bai F, Guddat LW, Yang H, Rao Z, Zhang B
Protein Cell (2023) 14 pp. 448-458 [ Pubmed: 36882106 DOI: doi:10.1093/procel/pwac060 ]
- Mycobactin import atp-binding/permease protein irta (93 kDa, Protein from Mycobacterium tuberculosis H37Rv)
- Mycobactin import atp-binding/permease protein irtb (61 kDa, Protein from Mycobacterium tuberculosis H37Rv)
- Phosphoaminophosphonic acid-adenylate ester (506 Da, Ligand)
- Irtab (Complex from Mycobacterium tuberculosis H37Rv)
Cryo-EM structure of EcmrR-RNAP-promoter open complex (EcmrR-RPo)
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 6xl5
Q-score: 0.541
Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, Hu Y, Liu B
Nat Commun (2021) 12 pp. 2702-2702 [ DOI: doi:10.1038/s41467-021-22990-8 Pubmed: 33976201 ]
- Ecmrr-rnap-promoter open complex (ecmrr-rpo) (Complex from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli O157:H7)
- Zinc ion (65 Da, Ligand)
- Water (18 Da, Ligand)
- Synthetic template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli O157:H7)
- Tetraphenylantimonium ion (430 Da, Ligand)
- Merr family transcriptional regulator ecmrr (31 kDa, Protein from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli O157:H7)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli O157:H7)
- Magnesium ion (24 Da, Ligand)
- Synthetic non-template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Chapso (631 Da, Ligand)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli O157:H7)
Resolution: 2.5 Å
EM Method: Single-particle
Fitted PDBs: 6xl9
Q-score: 0.517
Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, Hu Y, Liu B
Nat Commun (2021) 12 pp. 2702-2702 [ DOI: doi:10.1038/s41467-021-22990-8 Pubmed: 33976201 ]
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli O157:H7)
- Zinc ion (65 Da, Ligand)
- Water (18 Da, Ligand)
- Rna (5'-d(*(gtp))-r(p*ap*g)-3') (1 kDa, RNA from Escherichia coli O157:H7)
- Synthetic template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli O157:H7)
- Tetraphenylantimonium ion (430 Da, Ligand)
- Ecmrr-rnap-promoter initial transcribing complex with 3-nt rna transcript (ecmrr-rpitc-3nt) (Complex from Escherichia coli O157:H7)
- Merr family transcriptional regulator ecmrr (31 kDa, Protein from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli O157:H7)
- Magnesium ion (24 Da, Ligand)
- Synthetic non-template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli O157:H7)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6xlj
Q-score: 0.512
Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, Hu Y, Liu B
Nat Commun (2021) 12 pp. 2702-2702 [ DOI: doi:10.1038/s41467-021-22990-8 Pubmed: 33976201 ]
- Synthetic template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli O157:H7)
- Zinc ion (65 Da, Ligand)
- Ecmrr-rnap-promoter initial transcribing complex with 4-nt rna transcript (ecmrr-rpitc-4nt) (Complex from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli O157:H7)
- Tetraphenylantimonium ion (430 Da, Ligand)
- Merr family transcriptional regulator ecmrr (31 kDa, Protein from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli O157:H7)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli O157:H7)
- Rna (5'-d(*(gtp))-r(p*cp*ap*g)-3') (1 kDa, RNA from Escherichia coli O157:H7)
- Magnesium ion (24 Da, Ligand)
- Synthetic non-template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Chapso (631 Da, Ligand)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli O157:H7)
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 6xll
Q-score: 0.519
Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, Hu Y, Liu B
Nat Commun (2021) 12 pp. 2702-2702 [ DOI: doi:10.1038/s41467-021-22990-8 Pubmed: 33976201 ]
- Synthetic template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli O157:H7)
- Rna transcript (5-mer) (1 kDa, RNA from Escherichia coli O157:H7)
- Zinc ion (65 Da, Ligand)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli O157:H7)
- Rnap-promoter initial transcribing complex with 5-nt rna transcript (rpitc-5nt) (Complex from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli O157:H7)
- Rna polymerase sigma factor rpod (70 kDa, Protein from Escherichia coli O157:H7)
- Magnesium ion (24 Da, Ligand)
- Synthetic non-template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Chapso (631 Da, Ligand)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli O157:H7)
Cryo-EM structure of E. coli RNAP-DNA elongation complex 2 (RDe2) in EcmrR-dependent transcription
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 6xln
Q-score: 0.528
Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, Hu Y, Liu B
Nat Commun (2021) 12 pp. 2702-2702 [ DOI: doi:10.1038/s41467-021-22990-8 Pubmed: 33976201 ]
- Synthetic template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta' (155 kDa, Protein from Escherichia coli O157:H7)
- Zinc ion (65 Da, Ligand)
- 9-nt rna transcript (2 kDa, RNA from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit beta (150 kDa, Protein from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit alpha (36 kDa, Protein from Escherichia coli O157:H7)
- Magnesium ion (24 Da, Ligand)
- Synthetic non-template strand dna (54-mer) (16 kDa, DNA from Escherichia coli O157:H7)
- Chapso (631 Da, Ligand)
- E. coli rnap-dna elongation complex 2 (rde2) (Complex from Escherichia coli O157:H7)
- Dna-directed rna polymerase subunit omega (10 kDa, Protein from Escherichia coli O157:H7)
SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors in reduced conditions
Resolution: 2.95 Å
EM Method: Single-particle
Fitted PDBs: 7btf
Q-score: 0.569
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z
Science (2020) 368 pp. 779-782 [ Pubmed: 32277040 DOI: doi:10.1126/science.abb7498 ]
- Zinc ion (65 Da, Ligand)
- Rna-directed rna polymerase (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (9 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Structure of sars-cov-2 rna-dependent rna polymerase in complex with cofactors in reduced condition (155 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)