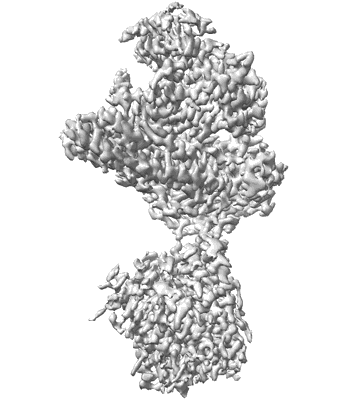
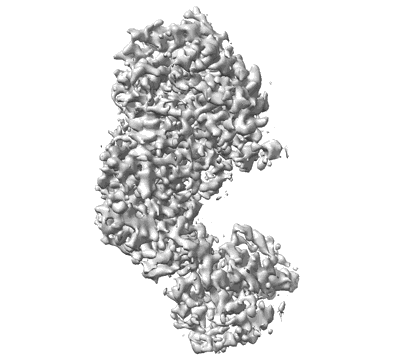Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8j5d
Q-score: 0.473
Liu J, Wang X, Guan Z, Wu M, Wang X, Fan R, Zhang F, Yan J, Liu Y, Zhang D, Yin P, Yan J
Plant Cell (2023) 36 pp. 194-212 [ DOI: doi:10.1093/plcell/koad259 Pubmed: 37804098 ]
- Beta-amylase 1, chloroplastic (56 kDa, Protein from Arabidopsis thaliana)
- Malate dehydrogenase, chloroplastic (34 kDa, Protein from Arabidopsis thaliana)
- Bam1-lsf1-mdh complex (Complex from Arabidopsis thaliana)
- Phosphoglucan phosphatase lsf1, chloroplastic (59 kDa, Protein from Arabidopsis thaliana)
Cryo-EM structure of VTC complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7ytj
Q-score: 0.485
Guan Z, Chen J, Liu R, Chen Y, Xing Q, Du Z, Cheng M, Hu J, Zhang W, Mei W, Wan B, Wang Q, Zhang J, Cheng P, Cai H, Cao J, Zhang D, Yan J, Yin P, Hothorn M, Liu Z
Nat Commun (2023) 14 pp. 718-718 [ DOI: doi:10.1038/s41467-023-36466-4 Pubmed: 36759618 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Vacuolar transporter chaperone 3 (100 kDa, Protein from Saccharomyces cerevisiae)
- Inositol hexakisphosphate (660 Da, Ligand)
- Vtc complex (Complex from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 4 (86 kDa, Protein from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 1 (15 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7yi8
Q-score: 0.499
Yan J, Liu F, Guan Z, Yan X, Jin X, Wang Q, Wang Z, Yan J, Zhang D, Liu Z, Wu S, Yin P
Cell Discov (2023) 9 pp. 8-8 [ DOI: doi:10.1038/s41421-022-00516-w Pubmed: 36658132 ]
- P1 (41 kDa, Protein from Tetrahymena thermophila SB210)
- Mta9 (52 kDa, Protein from Tetrahymena thermophila SB210)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Transmembrane protein, putative (19 kDa, Protein from Tetrahymena thermophila SB210)
- Mt-a70 family protein (42 kDa, Protein from Tetrahymena thermophila SB210)
- Mtac holoenzyme (Complex from Tetrahymena thermophila SB210)
Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 7yi9
Q-score: 0.522
Yan J, Liu F, Guan Z, Yan X, Jin X, Wang Q, Wang Z, Yan J, Zhang D, Liu Z, Wu S, Yin P
Cell Discov (2023) 9 pp. 8-8 [ DOI: doi:10.1038/s41421-022-00516-w Pubmed: 36658132 ]
- P1 (41 kDa, Protein from Tetrahymena thermophila SB210)
- Mta9 (52 kDa, Protein from Tetrahymena thermophila SB210)
- S-adenosylmethionine (398 Da, Ligand)
- Transmembrane protein, putative (19 kDa, Protein from Tetrahymena thermophila SB210)
- Mt-a70 family protein (42 kDa, Protein from Tetrahymena thermophila SB210)
- Mtac holoenzyme (Complex from Tetrahymena thermophila SB210)
Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7vgg
Q-score: 0.539
Wang Y, Wang L, Guan Z, Chang H, Ma L, Shen C, Qiu L, Yan J, Zhang D, Li J, Deng XW, Yin P
Sci Adv (2022) 8 pp. eabn3337-eabn3337 [ DOI: doi:10.1126/sciadv.abn3337 Pubmed: 35442727 ]
- Uvr8 (Complex)
- Cop1 (Complex from Arabidopsis thaliana)
- Uvr8-cop1 complex (Complex from Arabidopsis thaliana)
- Ultraviolet-b receptor uvr8 (50 kDa, Protein from Arabidopsis thaliana)
- E3 ubiquitin-protein ligase cop1 (79 kDa, Protein from Arabidopsis thaliana)
Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 7e4h
Q-score: 0.445
Wang Q, Guan Z, Qi L, Zhuang J, Wang C, Hong S, Yan L, Wu Y, Cao X, Cao J, Yan J, Zou T, Liu Z, Zhang D, Yan C, Yin P
Science (2021) 373 pp. 1377-1381 [ DOI: doi:10.1126/science.abh0704 Pubmed: 34446444 ]
- Sorting assembly machinery 50 kda subunit (54 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sam-tom40 complex (Complex from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery 35 kda subunit (37 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery 37 kda subunit (40 kDa, Protein from Saccharomyces cerevisiae S288c)
- Mitochondrial import receptor subunit tom40 (Complex)
- Mitochondrial import receptor subunit tom40 (46 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery subunits (Complex from Saccharomyces cerevisiae S288c)
Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom
Resolution: 3.05 Å
EM Method: Single-particle
Fitted PDBs: 7e4i
Q-score: 0.485
Wang Q, Guan Z, Qi L, Zhuang J, Wang C, Hong S, Yan L, Wu Y, Cao X, Cao J, Yan J, Zou T, Liu Z, Zhang D, Yan C, Yin P
Science (2021) 373 pp. 1377-1381 [ DOI: doi:10.1126/science.abh0704 Pubmed: 34446444 ]
- Mitochondrial import receptor subunit tom40 (43 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sam-tom40/tom5/tom6 complex (Complex from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery subunits (Complex from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery 50 kda subunit (54 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery 35 kda subunit (37 kDa, Protein from Saccharomyces cerevisiae S288c)
- Sorting assembly machinery 37 kda subunit (40 kDa, Protein from Saccharomyces cerevisiae S288c)
- Mitochondrial import receptor subunit tom5 (6 kDa, Protein from Saccharomyces cerevisiae S288c)
- Mitochondrial import receptor subunit tom6 (6 kDa, Protein from Saccharomyces cerevisiae S288c)
- Mitochondrial import receptor subunits (Complex)