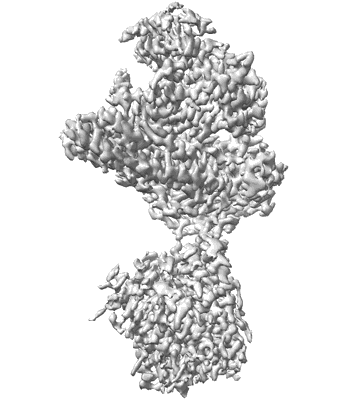
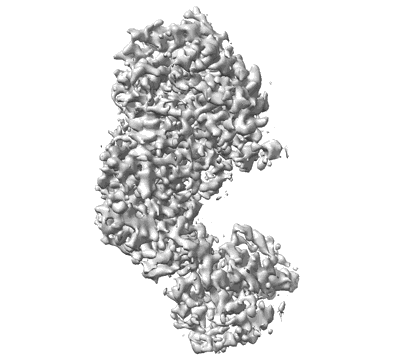CryoEM structure of Shedu from Bacillus cereus
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8ti8
Q-score: 0.532
Gu Y, Li H, Deep A, Enustun E, Zhang D, Corbett KD
Mol Cell (2025) 85 pp. 523-536.e6 [ DOI: doi:10.1016/j.molcel.2024.12.004 Pubmed: 39742666 ]
- Shedu protein sdua (44 kDa, Protein from Bacillus cereus B4264)
- Bacillus cereus shedu tetramer (175 kDa, Complex from Bacillus cereus B4264)
CryoEM structure of octamer assembly of Shedu nuclease domain from Bacillus cereus
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 8ti9
Q-score: 0.442
Gu Y, Li H, Deep A, Enustun E, Zhang D, Corbett KD
Mol Cell (2025) 85 pp. 523-536.e6 [ DOI: doi:10.1016/j.molcel.2024.12.004 Pubmed: 39742666 ]
- Shedu protein sdua (26 kDa, Protein from Bacillus cereus B4264)
- Bacillus cereus shedu delta_nl octamer (216 kDa, Complex from Bacillus cereus B4264)
Complex of NPR1 ectodomain with ANP plus an allosteric activating antibody, REGN5381
Resolution: 3.08 Å
EM Method: Single-particle
Fitted PDBs: 8tg9
Q-score: 0.495
Dunn ME, Kithcart A, Kim JH, Ho AJ, Franklin MC, Romero Hernandez A, de Hoon J, Botermans W, Meyer J, Jin X, Zhang D, Torello J, Jasewicz D, Kamat V, Garnova E, Liu N, Rosconi M, Pan H, Karnik S, Burczynski ME, Zheng W, Rafique A, Nielsen JB, De T, Verweij N, Pandit A, Locke A, Chalasani N, Melander O, Schwantes-An TH, Baras A, Lotta LA, Musser BJ, Mastaitis J, Devalaraja-Narashimha KB, Rankin AJ, Huang T, Herman G, Olson W, Murphy AJ, Yancopoulos GD, Olenchock BA, Morton L
Nature (2024) 633 pp. 654-661 [ DOI: doi:10.1038/s41586-024-07903-1 Pubmed: 39261724 ]
- Regn5381 fab heavy chain (24 kDa, Protein from Mus musculus)
- Atrial natriuretic peptide receptor 1 (52 kDa, Protein from Homo sapiens)
- Regn5381 fab light chain (23 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Active complex of npr1 ectodomain, anp, and regn5381 fab (Complex from Homo sapiens)
- Atrial natriuretic peptide (3 kDa, Protein from Homo sapiens)
Complex of NPR1 ectodomain and REGN5381 Fab in an active-like state with no ANP bound
Resolution: 3.65 Å
EM Method: Single-particle
Fitted PDBs: 8tga
Q-score: 0.117
Dunn ME, Kithcart A, Kim JH, Ho AJ, Franklin MC, Romero Hernandez A, de Hoon J, Botermans W, Meyer J, Jin X, Zhang D, Torello J, Jasewicz D, Kamat V, Garnova E, Liu N, Rosconi M, Pan H, Karnik S, Burczynski ME, Zheng W, Rafique A, Nielsen JB, De T, Verweij N, Pandit A, Locke A, Chalasani N, Melander O, Schwantes-An TH, Baras A, Lotta LA, Musser BJ, Mastaitis J, Devalaraja-Narashimha KB, Rankin AJ, Huang T, Herman G, Olson W, Murphy AJ, Yancopoulos GD, Olenchock BA, Morton L
Nature (2024) 633 pp. 654-661 [ DOI: doi:10.1038/s41586-024-07903-1 Pubmed: 39261724 ]
- Active-like complex of npr1 ectodomain and regn5381 fab, with no anp present (Complex from Homo sapiens)
- Regn5381 fab heavy chain (24 kDa, Protein from Mus musculus)
- Regn5381 fab light chain (23 kDa, Protein from Mus musculus)
- Atrial natriuretic peptide receptor 1 (52 kDa, Protein from Homo sapiens)
Resolution: 4.3 Å
EM Method: Single-particle
Fitted PDBs: 8w20
Q-score: 0.295
Zhao Q, Bertolli S, Park YJ, Tan Y, Cutler KJ, Srinivas P, Asfahl KL, Fonesca-Garcia C, Gallagher LA, Li Y, Wang Y, Coleman-Derr D, DiMaio F, Zhang D, Peterson SB, Veesler D, Mougous JD
Nature (2024) 629 pp. 165-173 [ Pubmed: 38632398 DOI: doi:10.1038/s41586-024-07298-z ]
- Umb1 umbrella toxin particle (Complex from Streptomyces coelicolor A3(2))
- Secreted protein (17 kDa, Protein from Streptomyces coelicolor A3(2))
- Intein c-terminal splicing domain-containing protein (142 kDa, Protein from Streptomyces coelicolor A3(2))
- Secreted esterase (54 kDa, Protein from Streptomyces coelicolor A3(2))
Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1)
Resolution: 4.0 Å
EM Method: Single-particle
Fitted PDBs: 8w22
Q-score: 0.386
Zhao Q, Bertolli S, Park YJ, Tan Y, Cutler KJ, Srinivas P, Asfahl KL, Fonesca-Garcia C, Gallagher LA, Li Y, Wang Y, Coleman-Derr D, DiMaio F, Zhang D, Peterson SB, Veesler D, Mougous JD
Nature (2024) 629 pp. 165-173 [ Pubmed: 38632398 DOI: doi:10.1038/s41586-024-07298-z ]
- Secreted protein (17 kDa, Protein from Streptomyces coelicolor A3(2))
- Umb1 umbrella toxin particle (local refinement of umbb1 bound alf of umbc1 and umba1) (Complex from Streptomyces coelicolor A3(2))
- Intein c-terminal splicing domain-containing protein (142 kDa, Protein from Streptomyces coelicolor A3(2))
- Secreted esterase (54 kDa, Protein from Streptomyces coelicolor A3(2))
Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2
Resolution: 2.98 Å
EM Method: Single-particle
Fitted PDBs: 8hfx
Q-score: 0.46
Han P, Meng Y, Zhang D, Xu Z, Li Z, Pan X, Zhao Z, Li L, Tang L, Qi J, Liu K, Gao GF
J Virol (2023) 97 pp. e0050523-e0050523 [ DOI: doi:10.1128/jvi.00505-23 Pubmed: 37676003 ]
- Spike glycoprotein,envelope glycoprotein (139 kDa, Protein from Human immunodeficiency virus 1)
- Angiotensin-converting enzyme (71 kDa, Protein from Odocoileus virginianus texanus)
- Zinc ion (65 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 omicron ba.1 spike protein, envelope glycoprotein chimera in complex with white-tailed deer ace2 (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of VTC complex
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 7ytj
Q-score: 0.485
Guan Z, Chen J, Liu R, Chen Y, Xing Q, Du Z, Cheng M, Hu J, Zhang W, Mei W, Wan B, Wang Q, Zhang J, Cheng P, Cai H, Cao J, Zhang D, Yan J, Yin P, Hothorn M, Liu Z
Nat Commun (2023) 14 pp. 718-718 [ DOI: doi:10.1038/s41467-023-36466-4 Pubmed: 36759618 ]
- Phosphate ion (94 Da, Ligand)
- 1,2-diacyl-sn-glycero-3-phosphocholine (790 Da, Ligand)
- Vacuolar transporter chaperone 3 (100 kDa, Protein from Saccharomyces cerevisiae)
- Inositol hexakisphosphate (660 Da, Ligand)
- Vtc complex (Complex from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 4 (86 kDa, Protein from Saccharomyces cerevisiae)
- Vacuolar transporter chaperone 1 (15 kDa, Protein from Saccharomyces cerevisiae)
Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2
Resolution: 2.66 Å
EM Method: Single-particle
Fitted PDBs: 7wrh
Q-score: 0.468
Li L, Han P, Huang B, Xie Y, Li W, Zhang D, Han P, Xu Z, Bai B, Zhou J, Kang X, Li X, Zheng A, Zhang R, Qiao S, Zhao X, Qi J, Wang Q, Liu K, Gao GF
Cell Discov (2022) 8 pp. 65-65 [ Pubmed: 35821014 DOI: doi:10.1038/s41421-022-00431-0 ]
- Mouse ace2 (Complex)
- Complex of sars-cov-2 omicron ba.1 spike protein with mouse ace2 (Complex from Severe acute respiratory syndrome coronavirus 2)
- Zinc ion (65 Da, Ligand)
- Processed angiotensin-converting enzyme 2 (69 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Spike glycoprotein (138 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron ba.1 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 7yi8
Q-score: 0.499
Yan J, Liu F, Guan Z, Yan X, Jin X, Wang Q, Wang Z, Yan J, Zhang D, Liu Z, Wu S, Yin P
Cell Discov (2023) 9 pp. 8-8 [ DOI: doi:10.1038/s41421-022-00516-w Pubmed: 36658132 ]
- P1 (41 kDa, Protein from Tetrahymena thermophila SB210)
- Mta9 (52 kDa, Protein from Tetrahymena thermophila SB210)
- S-adenosyl-l-homocysteine (384 Da, Ligand)
- Transmembrane protein, putative (19 kDa, Protein from Tetrahymena thermophila SB210)
- Mt-a70 family protein (42 kDa, Protein from Tetrahymena thermophila SB210)
- Mtac holoenzyme (Complex from Tetrahymena thermophila SB210)