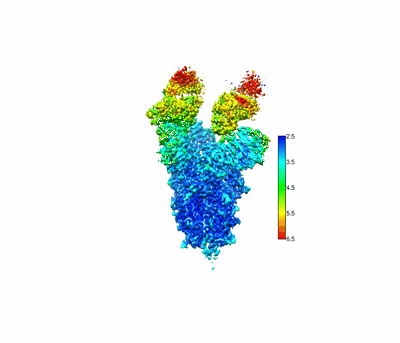
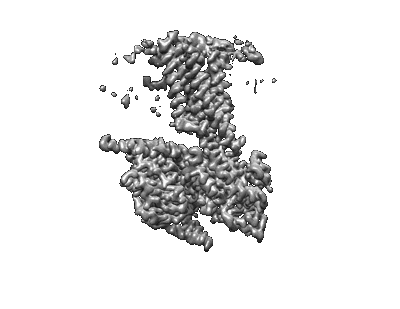SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
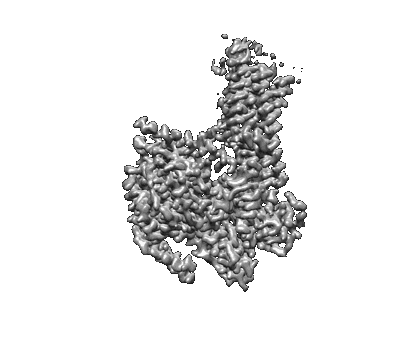
The interface structure of Omicron RBD binding to 5817 Fab
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8khd
Q-score: 0.393
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 (13 kDa, Protein from Homo sapiens)
- The interface structure of omicron rbd binding to 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Light chain of 5817 (11 kDa, Protein from Homo sapiens)
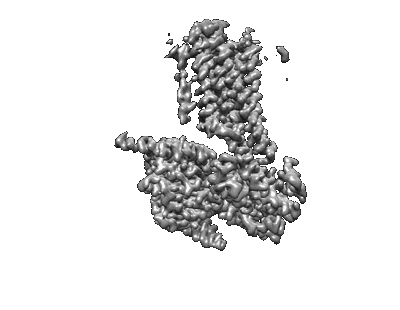
Cryo-EM structure of a class T GPCR in apo state
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 7xp4
Q-score: 0.428
Xu W, Wu L, Liu S, Liu X, Cao X, Zhou C, Zhang J, Fu Y, Guo Y, Wu Y, Tan Q, Wang L, Liu J, Jiang L, Fan Z, Pei Y, Yu J, Cheng J, Zhao S, Hao X, Liu ZJ, Hua T
Science (2022) 377 pp. 1298-1304 [ Pubmed: 36108005 DOI: doi:10.1126/science.abo1633 ]
- Guanine nucleotide-binding protein g(t) subunit alpha-3 (30 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Complex of gpcr and g protein (Complex from Homo sapiens)
- Nanobody 35 (15 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
- Endoglucanase h,taste receptor type 2 member 46,endoglucanase h,taste receptor type 2 member 46,bitter taste receptor t2r46 (91 kDa, Protein from Homo sapiens)
Cryo-EM structure of a class T GPCR in ligand-free state
Resolution: 3.08 Å
EM Method: Single-particle
Fitted PDBs: 7xp5
Q-score: 0.412
Xu W, Wu L, Liu S, Liu X, Cao X, Zhou C, Zhang J, Fu Y, Guo Y, Wu Y, Tan Q, Wang L, Liu J, Jiang L, Fan Z, Pei Y, Yu J, Cheng J, Zhao S, Hao X, Liu ZJ, Hua T
Science (2022) 377 pp. 1298-1304 [ Pubmed: 36108005 DOI: doi:10.1126/science.abo1633 ]
- Guanine nucleotide-binding protein g(t) subunit alpha-3 (30 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Endoglucanase h,taste receptor type 2 member 46,bitter taste receptor t2r46 (91 kDa, Protein from Homo sapiens)
- Complex of gpcr and g protein (Complex from Homo sapiens)
- Nanobody 35 (15 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
Cryo-EM structure of a class T GPCR in active state
Resolution: 3.01 Å
EM Method: Single-particle
Fitted PDBs: 7xp6
Q-score: 0.415
Xu W, Wu L, Liu S, Liu X, Cao X, Zhou C, Zhang J, Fu Y, Guo Y, Wu Y, Tan Q, Wang L, Liu J, Jiang L, Fan Z, Pei Y, Yu J, Cheng J, Zhao S, Hao X, Liu ZJ, Hua T
Science (2022) 377 pp. 1298-1304 [ Pubmed: 36108005 DOI: doi:10.1126/science.abo1633 ]
- Strychnine (334 Da, Ligand)
- Guanine nucleotide-binding protein g(t) subunit alpha-3 (30 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Complex of gpcr and g protein (Complex from Homo sapiens)
- Nanobody 35 (15 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
- Endoglucanase h,taste receptor type 2 member 46,endoglucanase h,taste receptor type 2 member 46,bitter taste receptor t2r46 (91 kDa, Protein from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement)
Resolution: 3.88 Å
EM Method: Single-particle
Fitted PDBs: 7xrp
Q-score: 0.447
Zhao D, Liu L, Liu X, Zhang J, Yin Y, Luan L, Jiang D, Yang X, Li L, Xiong H, Xing D, Zheng Q, Xia N, Tao Y, Li S, Huang H
J Nanobiotechnology (2022) 20 pp. 411-411 [ Pubmed: 36109732 DOI: doi:10.1186/s12951-022-01619-y ]
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Nanobody c5g2 (Complex)
- Spike protein s1 (32 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike protein (Complex from Camelus dromedarius)
- C5g2 nanobody (13 kDa, Protein from Camelus dromedarius)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Cryo-em structure of sars-cov-2 spike protein in complex with nanobody c5g2 (local refinement) (Complex from Severe acute respiratory syndrome coronavirus 2)
2-fold subparticles refinement of human papillomavirus type 58 pseudovirus
3-fold sub-particles refinement of human papillomavirus type 58 pseudovirus
5-fold sub-particles refinement of human papillomavirus type 58 pseudovirus