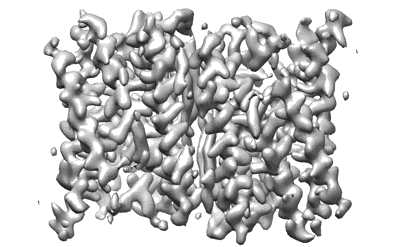
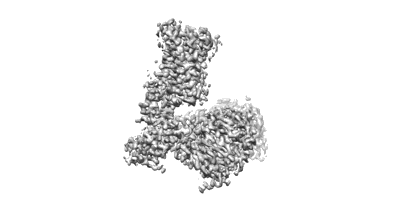
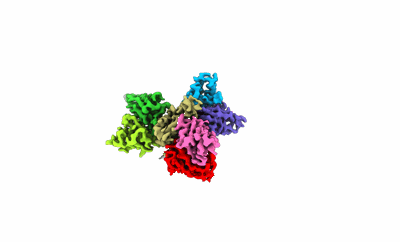
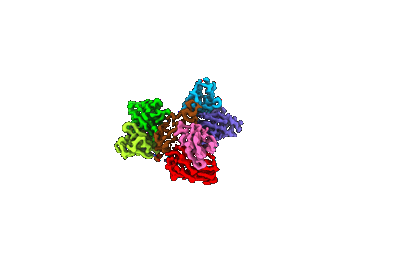
The structure of EfpA_BRD-8000.3 complex
Resolution: 3.41 Å
EM Method: Single-particle
Fitted PDBs: 8ynz
Q-score: 0.406
Li D, Zhang X, Yao Y, Sun X, Sun J, Ma X, Yuan K, Bai G, Pang X, Hua R, Guo T, Mi Y, Wu L, Zhang J, Wu Y, Liu Y, Wang P, Wong CCL, Chen XW, Xiao H, Gao GF, Gao F
PNAS (2024) 121 pp. e2412653121-e2412653121 [ DOI: doi:10.1073/pnas.2412653121 Pubmed: 39441632 ]
- Efpa (Complex from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv))
- (1s,3s)-n-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3-(2-methylprop-1-en-1-yl)cyclopropane-1-carboxamide (401 Da, Ligand)
- Uncharacterized mfs-type transporter efpa (59 kDa, Protein from Mycobacterium tuberculosis H37Rv)
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
Cryo-EM map of Gi-scFv16 complex
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Resolution: 3.34 Å
EM Method: Single-particle
Fitted PDBs: 8iyx
Q-score: 0.448
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
- 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid (583 Da, Ligand)
- Probable g-protein coupled receptor 34,probable g-protein coupled receptor 34,yl-365 (66 kDa, Protein from Homo sapiens)
- Cryo-em structure of the gpr34 receptor in complex with the antagonist yl-365 (Complex from Homo sapiens)
Cryo-EM structure of GPR34-Gi complex
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8sai
Q-score: 0.471
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
- Scfv16 (Complex from Mus musculus)
- Human gpr34 in complex with gi heterotrimer and scfv16 (Complex)
- O-[(s)-hydroxy{[(2s)-2-hydroxy-3-(octadec-9-enoyloxy)propyl]oxy}phosphoryl]-l-serine (523 Da, Ligand)
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Probable g-protein coupled receptor 34 (53 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Scfv16 (28 kDa, Protein from Mus musculus)
- Soluble cytochrome b562,probable g-protein coupled receptor 34, guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2, guanine nucleotide-binding protein g(i) subunit alpha-1, guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (Complex from Homo sapiens)
Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17
Resolution: 3.48 Å
EM Method: Single-particle
Fitted PDBs: 7x7t
Q-score: 0.504
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- Sars-cov-2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- Cryo-em structure of sars-cov-2 spike protein in complex with three nabs x01, x10 and x17 (Complex)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- X10 light chain (12 kDa, Protein from Mus musculus)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- X01 heavy chain (13 kDa, Protein from Mus musculus)
Resolution: 3.77 Å
EM Method: Single-particle
Fitted PDBs: 7x7u
Q-score: 0.49
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- Cryo-em structure of sars-cov-2 delta variant spike protein in complex with three nabs x01, x10 and x17 (Complex)
- Spike protein s1 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 spike protein (Complex from Severe acute respiratory syndrome coronavirus 2)
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- X10 light chain (12 kDa, Protein from Mus musculus)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- X01 heavy chain (13 kDa, Protein from Mus musculus)
Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17
Resolution: 3.83 Å
EM Method: Single-particle
Fitted PDBs: 7x7v
Q-score: 0.484
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]
- Nabs x01, x10 and x17 (Complex from Mus musculus)
- X17 heavy chain (13 kDa, Protein from Mus musculus)
- X01 light chain (11 kDa, Protein from Mus musculus)
- Cryo-em structure of sars-cov spike protein in complex with three nabs x01, x10 and x17 (Complex)
- Spike protein s1 (21 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- X10 light chain (12 kDa, Protein from Mus musculus)
- X17 light chain (11 kDa, Protein from Mus musculus)
- X10 heavy chain (13 kDa, Protein from Mus musculus)
- X01 heavy chain (13 kDa, Protein from Mus musculus)
- Sars-cov spike protein (Complex from Severe acute respiratory syndrome coronavirus)
Xiong H, Sun H, Wang S, Yuan L, Liu L, Zhu Y, Zhang J, Huang Y, Qi R, Jiang Y, Ma J, Zhou M, Ma Y, Fu R, Yan S, Yue M, Wu Y, Wei M, Wang Y, Li T, Wang Y, Zheng Z, Yu H, Cheng T, Li S, Yuan Q, Zhang J, Guan Y, Zheng Q, Zhang T, Xia N
PNAS (2022) 119 pp. e2204256119-e2204256119 [ DOI: doi:10.1073/pnas.2204256119 Pubmed: 35972965 ]