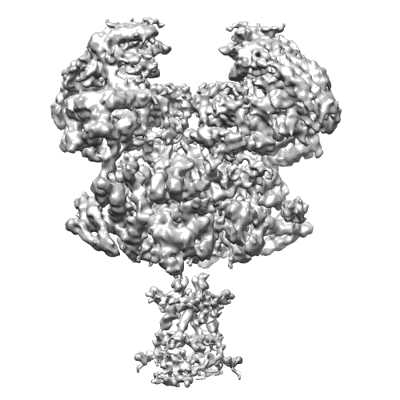
Cryo-EM structure of 5-subunit Smc5/6 arm region
Resolution: 5.97 Å
EM Method: Single-particle
Fitted PDBs: 8i4v
Q-score: 0.108
Li Q, Zhang J, Haluska C, Zhang X, Wang L, Liu G, Wang Z, Jin D, Cheng T, Wang H, Tian Y, Wang X, Sun L, Zhao X, Chen Z, Wang L
Nat Struct Mol Biol (2024) 31 pp. 1532-1542 [ Pubmed: 38890552 DOI: doi:10.1038/s41594-024-01319-1 ]
- Dna repair protein kre29 (7 kDa, Protein from Saccharomyces cerevisiae S288C)
- Structural maintenance of chromosomes protein 5 (123 kDa, Protein from Saccharomyces cerevisiae S288C)
- E3 sumo-protein ligase mms21 (28 kDa, Protein from Saccharomyces cerevisiae S288C)
- Cryo-em structure of 5-subunit smc5/6 arm region (Complex from Saccharomyces cerevisiae)
- Structural maintenance of chromosomes protein 6 (128 kDa, Protein from Saccharomyces cerevisiae S288C)
Cryo-EM structure of 6-subunit Smc5/6 head region
Resolution: 5.58 Å
EM Method: Single-particle
Fitted PDBs: 8wjn
Q-score: 0.198
Li Q, Zhang J, Haluska C, Zhang X, Wang L, Liu G, Wang Z, Jin D, Cheng T, Wang H, Tian Y, Wang X, Sun L, Zhao X, Chen Z, Wang L
Nat Struct Mol Biol (2024) 31 pp. 1532-1542 [ Pubmed: 38890552 DOI: doi:10.1038/s41594-024-01319-1 ]
- Cryo-em structure of 6-subunit smc5/6 head region (Complex from Saccharomyces cerevisiae)
- Non-structural maintenance of chromosomes element 4 (46 kDa, Protein from Saccharomyces cerevisiae S288C)
- Structural maintenance of chromosomes protein 6 (128 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosomes element 1 (38 kDa, Protein from Saccharomyces cerevisiae S288C)
- Structural maintenance of chromosomes protein 5 (126 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosome element 3 (34 kDa, Protein from Saccharomyces cerevisiae S288C)
Cryo-EM structure of 8-subunit Smc5/6
Resolution: 5.6 Å
EM Method: Single-particle
Fitted PDBs: 7yqh
Q-score: 0.19
Li Q, Zhang J, Haluska C, Zhang X, Wang L, Liu G, Wang Z, Jin D, Cheng T, Wang H, Tian Y, Wang X, Sun L, Zhao X, Chen Z, Wang L
Nat Struct Mol Biol (2024) [ Pubmed: 38890552 DOI: doi:10.1038/s41594-024-01319-1 ]
- E3 sumo-protein ligase mms21 (30 kDa, Protein from Saccharomyces cerevisiae S288C)
- Dna repair protein kre29 (53 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosomes element 4 (46 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosome element 5 (64 kDa, Protein from Saccharomyces cerevisiae S288C)
- Cryo-em structure of 8-subunit smc5/6 (Complex from Saccharomyces cerevisiae S288C)
- Structural maintenance of chromosomes protein 6 (128 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosomes element 1 (38 kDa, Protein from Saccharomyces cerevisiae S288C)
- Structural maintenance of chromosomes protein 5 (126 kDa, Protein from Saccharomyces cerevisiae S288C)
- Non-structural maintenance of chromosome element 3 (34 kDa, Protein from Saccharomyces cerevisiae S288C)
Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state.
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 7yfr
Q-score: 0.388
Zhang J, Zhang M, Wang Q, Wen H, Liu Z, Wang F, Wang Y, Yao F, Song N, Kou Z, Li Y, Guo F, Zhu S
Nat Struct Mol Biol (2023) 30 pp. 629-639 [ Pubmed: 36959261 DOI: doi:10.1038/s41594-023-00959-z ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Glutamate receptor ionotropic, nmda 2d (97 kDa, Protein from Homo sapiens)
- Glutamic acid (147 Da, Ligand)
- Nmda receptor with nmda 1 incorperated with nmda 2d (384 MDa, Complex from Homo sapiens)
- Glycine (75 Da, Ligand)
- Glutamate receptor ionotropic, nmda 1 (95 kDa, Protein from Homo sapiens)
Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate.
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 7yfm
Q-score: 0.309
Zhang J, Zhang M, Wang Q, Wen H, Liu Z, Wang F, Wang Y, Yao F, Song N, Kou Z, Li Y, Guo F, Zhu S
Nat Struct Mol Biol (2023) 30 pp. 629-639 [ Pubmed: 36959261 DOI: doi:10.1038/s41594-023-00959-z ]
- Isoform 6 of glutamate receptor ionotropic, nmda 1 (97 kDa, Protein from Homo sapiens)
- Glutamate receptor ionotropic, nmda 2d (97 kDa, Protein from Homo sapiens)
- Nmda receptor with nmda 1 incorperated with nmda 2d (384 MDa, Complex from Homo sapiens)
The atomic structure of varicella zoster virus C-capsid
Wang W, Zheng Q, Pan D, Yu H, Fu W, Liu J, He M, Zhu R, Cai Y, Huang Y, Zha Z, Chen Z, Ye X, Han J, Que Y, Wu T, Zhang J, Li S, Zhu H, Zhou ZH, Cheng T, Xia N
Nat Microbiol (2020) 5 pp. 1542-1552 [ DOI: doi:10.1038/s41564-020-0785-y Pubmed: 32895526 ]
- Major capsid protein (155 kDa, Protein from Human herpesvirus 3)
- Human alphaherpesvirus 3 (Virus from Human alphaherpesvirus 3)
- Triplex capsid protein 1 (54 kDa, Protein from Human herpesvirus 3)
- Small capsomere-interacting protein (24 kDa, Protein from Human herpesvirus 3)
- Triplex capsid protein 2 (34 kDa, Protein from Human herpesvirus 3)
Resolution: 5.1 Å
EM Method: Single-particle
Fitted PDBs: 6irf
Q-score: 0.158
Resolution: 5.5 Å
EM Method: Single-particle
Fitted PDBs: 6irg
Q-score: 0.147
Cryo-EM map of F.graminearum Mitochondrial Calcium Uniporter in PMAL-C8
Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L
Nature (2018) 559 pp. 575-579 [ Pubmed: 29995856 DOI: doi:10.1038/s41586-018-0330-9 ]