ACAD11 D220A with 4-hydroxyvaleryl-CoA
Resolution: 2.6 Å
EM Method: Single-particle
Fitted PDBs: 8v3u
Q-score: 0.637
ACAD11 D753N with 4-phosphovaleryl-CoA
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 8v3v
Q-score: 0.471
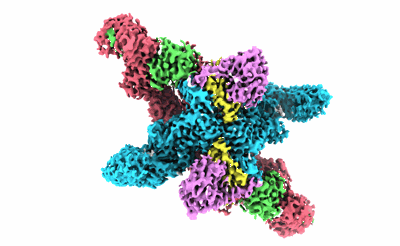
The CryoEM structure of the high affinity Carbon monoxide dehydrogenase from Mycobacterium smegmatis
Resolution: 1.85 Å
EM Method: Single-particle
Fitted PDBs: 8uem
Q-score: 0.777
Kropp A, Gillett DL, Venugopal H, Gonzalvez MA, Lingford JP, Jain S, Barlow CK, Zhang J, Greening C, Grinter R
Nat Chem Biol (2025) [ DOI: doi:10.1038/s41589-025-01836-0 Pubmed: 39881213 ]
- [2fe-2s] binding domain protein (17 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Flavin-adenine dinucleotide (785 Da, Ligand)
- Fe2/s2 (inorganic) cluster (175 Da, Ligand)
- Carbon monoxide dehydrogenase (large chain), coxl (85 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Cu(i)-s-mo(iv)(=o)oh cluster (224 Da, Ligand)
- Carbon monoxide dehydrogenase medium chain (31 kDa, Protein from Mycolicibacterium smegmatis MC2 155)
- Mocu carbon monoxide dehydrogenase (mocu-codh) (270 kDa, Complex from Mycolicibacterium smegmatis MC2 155)
- Water (18 Da, Ligand)
- Pterin cytosine dinucleotide (696 Da, Ligand)
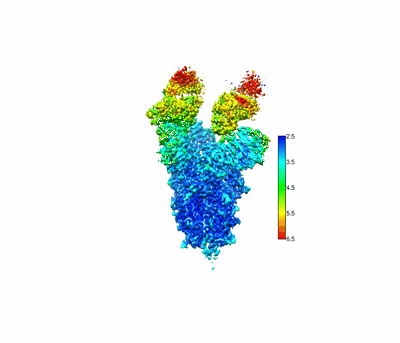
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ Pubmed: 38175758 DOI: doi:10.1016/j.celrep.2023.113653 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
The interface structure of Omicron RBD binding to 5817 Fab
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8khd
Q-score: 0.393
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ Pubmed: 38175758 DOI: doi:10.1016/j.celrep.2023.113653 ]
- Light chain of 5817 (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- The interface structure of omicron rbd binding to 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 (13 kDa, Protein from Homo sapiens)
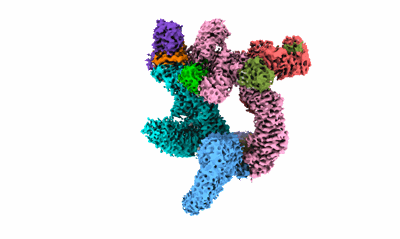
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1)
Resolution: 3.39 Å
EM Method: Single-particle
Fitted PDBs: 8wqa
Q-score: 0.362
Chen X, Raiff A, Li S, Guo Q, Zhang J, Zhou H, Timms RT, Yao X, Elledge SJ, Koren I, Zhang K, Xu C
Nat Commun (2024) 15 pp. 3558-3558 [ Pubmed: 38670995 DOI: doi:10.1038/s41467-024-47890-5 ]
- Cryo-em structure of cul2-rbx1-elob-eloc-fem1b bound with the c-degron of ccdc89 (conformation 1) (Complex from Homo sapiens)
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (11 kDa, Protein from Homo sapiens)
- Protein fem-1 homolog b (70 kDa, Protein from Homo sapiens)
- Elongin-c (10 kDa, Protein from Homo sapiens)
- Cullin-2 (87 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Coiled-coil domain-containing protein 89 (3 kDa, Protein from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2)
Resolution: 3.37 Å
EM Method: Single-particle
Fitted PDBs: 8wqb
Q-score: 0.39
Chen X, Raiff A, Li S, Guo Q, Zhang J, Zhou H, Timms RT, Yao X, Elledge SJ, Koren I, Zhang K, Xu C
Nat Commun (2024) 15 pp. 3558-3558 [ Pubmed: 38670995 DOI: doi:10.1038/s41467-024-47890-5 ]
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (11 kDa, Protein from Homo sapiens)
- Protein fem-1 homolog b (70 kDa, Protein from Homo sapiens)
- Elongin-c (10 kDa, Protein from Homo sapiens)
- Cullin-2 (87 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Cryo-em structure of cul2-rbx1-elob-eloc-fem1b bound with the c-degron of ccdc89 (conformation 2) (Complex from Homo sapiens)
- Coiled-coil domain-containing protein 89 (3 kDa, Protein from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)
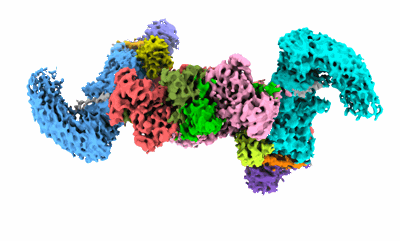
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1
Resolution: 3.54 Å
EM Method: Single-particle
Fitted PDBs: 8wqc
Q-score: 0.352
Chen X, Raiff A, Li S, Guo Q, Zhang J, Zhou H, Timms RT, Yao X, Elledge SJ, Koren I, Zhang K, Xu C
Nat Commun (2024) 15 pp. 3558-3558 [ Pubmed: 38670995 DOI: doi:10.1038/s41467-024-47890-5 ]
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (11 kDa, Protein from Homo sapiens)
- Protein fem-1 homolog b (70 kDa, Protein from Homo sapiens)
- Elongin-c (10 kDa, Protein from Homo sapiens)
- Cullin-2 (87 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Cryo-em structure of neddylated cul2-rbx1-elob-eloc-fem1b bound with the c-degron of cdk5r1 (Complex from Homo sapiens)
- Nedd8 (8 kDa, Protein from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1)
Resolution: 3.38 Å
EM Method: Single-particle
Fitted PDBs: 8wqe
Q-score: 0.372
Chen X, Raiff A, Li S, Guo Q, Zhang J, Zhou H, Timms RT, Yao X, Elledge SJ, Koren I, Zhang K, Xu C
Nat Commun (2024) 15 pp. 3558-3558 [ Pubmed: 38670995 DOI: doi:10.1038/s41467-024-47890-5 ]
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (11 kDa, Protein from Homo sapiens)
- Protein fem-1 homolog b (70 kDa, Protein from Homo sapiens)
- Elongin-c (10 kDa, Protein from Homo sapiens)
- Cullin-2 (87 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Protein casp (3 kDa, Protein from Homo sapiens)
- Cryo-em structure of cul2-rbx1-elob-eloc-fem1b bound with the c-degron of cux1 (conformation 1) (Complex from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)
cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2)
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8wqf
Q-score: 0.439
Chen X, Raiff A, Li S, Guo Q, Zhang J, Zhou H, Timms RT, Yao X, Elledge SJ, Koren I, Zhang K, Xu C
Nat Commun (2024) 15 pp. 3558-3558 [ Pubmed: 38670995 DOI: doi:10.1038/s41467-024-47890-5 ]
- E3 ubiquitin-protein ligase rbx1, n-terminally processed (11 kDa, Protein from Homo sapiens)
- Protein fem-1 homolog b (70 kDa, Protein from Homo sapiens)
- Cryo-em structure of cul2-rbx1-elob-eloc-fem1b bound with the c-degron of cux1 (conformation 2) (Complex from Homo sapiens)
- Elongin-c (10 kDa, Protein from Homo sapiens)
- Cullin-2 (87 kDa, Protein from Homo sapiens)
- Zinc ion (65 Da, Ligand)
- Protein casp (3 kDa, Protein from Homo sapiens)
- Elongin-b (13 kDa, Protein from Homo sapiens)