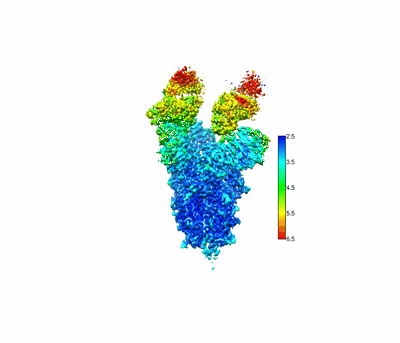
CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 8xko
Q-score: 0.486
Jia X, Jing X, Li M, Gao M, Zhong Y, Li E, Liu Y, Li R, Yao G, Liu Q, Zhou M, Hou Y, An L, Hong Y, Li S, Zhang J, Wang W, Zhang K, Gong P, Chiu S
Nat Commun (2024) 15 pp. 10750-10750 [ Pubmed: 39737930 DOI: doi:10.1038/s41467-024-54918-3 ]
- [[(2~{r},3~{r},4~{s},5~{r})-4-fluoranyl-5-(5-iodanyl-4-methyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate (633 Da, Ligand)
- Rna (5'-r(p*up*up*cp*ap*up*gp*cp*up*ap*cp*gp*cp*gp*up*ap*g)-3') (5 kDa, RNA from synthetic construct)
- Non-structural protein 8 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Magnesium ion (24 Da, Ligand)
- Rna (5'-r(p*cp*up*ap*cp*gp*cp*gp*up*ap*gp*cp*ap*up*g)-3') (4 kDa, RNA from synthetic construct)
- Cryoem structure of compound hnc-1644 bound with rdrp-rna (150 kDa, Complex from Severe acute respiratory syndrome coronavirus)
- Rna-directed rna polymerase nsp12 (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus)
Cryo-EM structure of Tdk1-Bdf1 complex
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 9ja5
Q-score: 0.524
Cryo-EM structure of noradrenaline transporter in apo state
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8wtu
Q-score: 0.614
Hu T, Yu Z, Zhao J, Meng Y, Salomon K, Bai Q, Wei Y, Zhang J, Xu S, Dai Q, Yu R, Yang B, Loland CJ, Zhao Y
Nature (2024) 632 pp. 930-937 [ DOI: doi:10.1038/s41586-024-07638-z Pubmed: 39085602 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Structure of a protein (63 MDa, Complex from Homo sapiens)
- Sodium-dependent noradrenaline transporter (63 kDa, Protein from Homo sapiens)
Cryo-EM structure of noradrenaline transporter in complex with noradrenaline
Resolution: 2.7 Å
EM Method: Single-particle
Fitted PDBs: 8wtv
Q-score: 0.655
Hu T, Yu Z, Zhao J, Meng Y, Salomon K, Bai Q, Wei Y, Zhang J, Xu S, Dai Q, Yu R, Yang B, Loland CJ, Zhao Y
Nature (2024) 632 pp. 930-937 [ DOI: doi:10.1038/s41586-024-07638-z Pubmed: 39085602 ]
- Noradrenaline (170 Da, Ligand)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Structure of a protein (63 MDa, Complex from Homo sapiens)
- Sodium-dependent noradrenaline transporter (63 kDa, Protein from Homo sapiens)
- Sodium ion (22 Da, Ligand)
- Chloride ion (35 Da, Ligand)
- Water (18 Da, Ligand)
Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8wtw
Q-score: 0.617
Hu T, Yu Z, Zhao J, Meng Y, Salomon K, Bai Q, Wei Y, Zhang J, Xu S, Dai Q, Yu R, Yang B, Loland CJ, Zhao Y
Nature (2024) 632 pp. 930-937 [ DOI: doi:10.1038/s41586-024-07638-z Pubmed: 39085602 ]
- Apo (Complex from Homo sapiens)
- Sodium-dependent noradrenaline transporter (63 kDa, Protein from Homo sapiens)
- Sodium ion (22 Da, Ligand)
- Mrla (1 kDa, Protein from Conus marmoreus)
- Chloride ion (35 Da, Ligand)
Structure of L797591-SSTR1 G protein complex
Resolution: 2.65 Å
EM Method: Single-particle
Fitted PDBs: 8xio
Q-score: 0.469
Wang Y, Xu Y, Wang Y, Zhang J, Chen L, He X, Fan W, Wu K, Hu W, Cheng X, Yang G, Xu HE, Zhuang Y, Sun S
PNAS (2024) 121 pp. e2400298121-e2400298121 [ DOI: doi:10.1073/pnas.2400298121 Pubmed: 39361640 ]
- (2~{s})-~{n}-[(4~{s})-6-azanyl-2,2,4-trimethyl-hexyl]-3-naphthalen-1-yl-2-[[2-phenylethyl(2-pyridin-2-ylethyl)carbamoyl]amino]propanamide (607 Da, Ligand)
- Somatostatin receptor type 1 (42 kDa, Protein from Homo sapiens)
- L797591-sstr1 g protein complex (160 kDa, Complex from Homo sapiens)
- G-alpha-i (41 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Nanobody nb35 (16 kDa, Protein from Lama glama)
- Water (18 Da, Ligand)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (38 kDa, Protein from Homo sapiens)
Structure of L796778-SSTR3 G protein complex
Resolution: 2.71 Å
EM Method: Single-particle
Fitted PDBs: 8xiq
Q-score: 0.462
Wang Y, Xu Y, Wang Y, Zhang J, Chen L, He X, Fan W, Wu K, Hu W, Cheng X, Yang G, Xu HE, Zhuang Y, Sun S
PNAS (2024) 121 pp. e2400298121-e2400298121 [ DOI: doi:10.1073/pnas.2400298121 Pubmed: 39361640 ]
- Structure of l796778-sstr3 g protein complex (160 kDa, Complex from Homo sapiens)
- Water (18 Da, Ligand)
- G-alpha i (41 kDa, Protein from Homo sapiens)
- Somatostatin receptor type 3 (40 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Nanobody nb35 (16 kDa, Protein from Lama glama)
- Nit-fc0-nle-a1d5b (584 Da, Protein from synthetic construct)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (40 kDa, Protein from Homo sapiens)
Structure of pasireotide-SSTR3 G protein complex
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8xir
Q-score: 0.501
Wang Y, Xu Y, Wang Y, Zhang J, Chen L, He X, Fan W, Wu K, Hu W, Cheng X, Yang G, Xu HE, Zhuang Y, Sun S
PNAS (2024) 121 pp. e2400298121-e2400298121 [ DOI: doi:10.1073/pnas.2400298121 Pubmed: 39361640 ]
- Somatostatin receptor type 3 (45 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (41 kDa, Protein from Homo sapiens)
- G-alpha i (41 kDa, Protein from Homo sapiens)
- Nanobody nb35 (16 kDa, Protein from Lama glama)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Pasireotide (1 kDa, Protein from synthetic construct)
- Pasireotide-sstr3 g protein complex (160 kDa, Complex from Homo sapiens)
- Water (18 Da, Ligand)
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ Pubmed: 38175758 DOI: doi:10.1016/j.celrep.2023.113653 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Water (18 Da, Ligand)
The interface structure of Omicron RBD binding to 5817 Fab
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8khd
Q-score: 0.393
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ Pubmed: 38175758 DOI: doi:10.1016/j.celrep.2023.113653 ]
- Light chain of 5817 (11 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- The interface structure of omicron rbd binding to 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 (13 kDa, Protein from Homo sapiens)