CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2
Resolution: 3.29 Å
EM Method: Single-particle
Fitted PDBs: 8xko
Q-score: 0.486
Jia X, Jing X, Li M, Gao M, Zhong Y, Li E, Liu Y, Li R, Yao G, Liu Q, Zhou M, Hou Y, An L, Hong Y, Li S, Zhang J, Wang W, Zhang K, Gong P, Chiu S
Nat Commun (2024) 15 pp. 10750-10750 [ Pubmed: 39737930 DOI: doi:10.1038/s41467-024-54918-3 ]
- Rna (5'-r(p*up*up*cp*ap*up*gp*cp*up*ap*cp*gp*cp*gp*up*ap*g)-3') (5 kDa, RNA from synthetic construct)
- Magnesium ion (24 Da, Ligand)
- Non-structural protein 7 (10 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- Rna (5'-r(p*cp*up*ap*cp*gp*cp*gp*up*ap*gp*cp*ap*up*g)-3') (4 kDa, RNA from synthetic construct)
- Cryoem structure of compound hnc-1644 bound with rdrp-rna (150 kDa, Complex from Severe acute respiratory syndrome coronavirus)
- [[(2~{r},3~{r},4~{s},5~{r})-4-fluoranyl-5-(5-iodanyl-4-methyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate (633 Da, Ligand)
- Rna-directed rna polymerase nsp12 (108 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Non-structural protein 8 (23 kDa, Protein from Severe acute respiratory syndrome coronavirus)
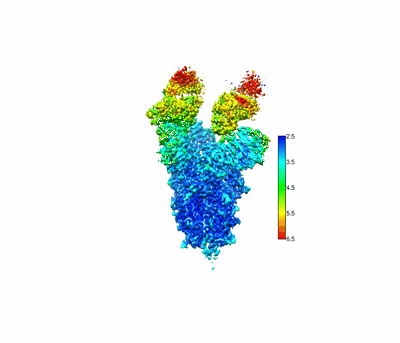
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
The interface structure of Omicron RBD binding to 5817 Fab
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 8khd
Q-score: 0.393
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Spike protein s1 (22 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 (13 kDa, Protein from Homo sapiens)
- The interface structure of omicron rbd binding to 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Light chain of 5817 (11 kDa, Protein from Homo sapiens)
Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8kd3
Q-score: 0.376
Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J
Cell Res (2023) 33 pp. 790-801 [ DOI: doi:10.1038/s41422-023-00869-1 Pubmed: 37666978 ]
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- H3k9 deacetylation state of rpd3s-ncp (187bp/mla/k9q) (Complex)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3-sin3-eaf3-rco1 (Complex from Saccharomyces cerevisiae)
- Histone (Complex from Xenopus laevis)
- Transcriptional regulatory protein rco1 (84 kDa, Protein from Saccharomyces cerevisiae)
- 187bp dna (58 kDa, DNA from synthetic construct)
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- 187bp dna (57 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Transcriptional regulatory protein sin3 (176 kDa, Protein from Saccharomyces cerevisiae)
- Dna (Complex)
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1
Resolution: 2.93 Å
EM Method: Single-particle
Fitted PDBs: 8kd4
Q-score: 0.373
Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J
Cell Res (2023) 33 pp. 790-801 [ DOI: doi:10.1038/s41422-023-00869-1 Pubmed: 37666978 ]
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- H3/h4 deacetylation state of rpd3s-ncp(187bp/mla)class1 (Complex)
- Histone (Complex from Xenopus laevis)
- Transcriptional regulatory protein rco1 (84 kDa, Protein from Saccharomyces cerevisiae)
- 187bp dna (58 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (157 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- 187bp dna (57 kDa, DNA from synthetic construct)
- Rpd3-sin3-rco1-eaf3 (Complex from Saccharomyces cerevisiae)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Dna (Complex)
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8kd5
Q-score: 0.378
Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J
Cell Res (2023) 33 pp. 790-801 [ DOI: doi:10.1038/s41422-023-00869-1 Pubmed: 37666978 ]
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3-sin3-eaf3-rco1 (Complex from Saccharomyces cerevisiae)
- Transcriptional regulatory protein rco1 (78 kDa, Protein from Saccharomyces cerevisiae)
- 187bp dna (Complex from synthetic construct)
- 187bp dna (58 kDa, DNA from synthetic construct)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- 187bp dna (57 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- Transcriptional regulatory protein sin3 (176 kDa, Protein from Saccharomyces cerevisiae)
- H3/h4 deacetylation state of rpd3s-ncp (187bp/mla) class2 (Complex)
- Histone (Complex from Xenopus laevis)
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3
Resolution: 3.07 Å
EM Method: Single-particle
Fitted PDBs: 8kd6
Q-score: 0.311
Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J
Cell Res (2023) 33 pp. 790-801 [ DOI: doi:10.1038/s41422-023-00869-1 Pubmed: 37666978 ]
- Histone h3 (15 kDa, Protein from Xenopus laevis)
- Histone deacetylase rpd3 (48 kDa, Protein from Saccharomyces cerevisiae)
- Rpd3-sin3-eaf3-rco1 (Complex from Saccharomyces cerevisiae)
- Histone (Complex from Xenopus laevis)
- Transcriptional regulatory protein rco1 (84 kDa, Protein from Saccharomyces cerevisiae)
- 187bp dna (58 kDa, DNA from synthetic construct)
- Transcriptional regulatory protein sin3 (157 kDa, Protein from Saccharomyces cerevisiae)
- Zinc ion (65 Da, Ligand)
- Histone h2a (13 kDa, Protein from Xenopus laevis)
- Histone h2b 1.1 (13 kDa, Protein from Xenopus laevis)
- 187bp dna (57 kDa, DNA from synthetic construct)
- Histone h4 (11 kDa, Protein from Xenopus laevis)
- Chromatin modification-related protein eaf3 (45 kDa, Protein from Saccharomyces cerevisiae)
- H3/h4 deacetylation state of rpd3s-ncp(187bp/mla) class3 (Complex)
- Dna (Complex)
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 7y71
Q-score: 0.415
Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Kwek MSS, Greaney AJ, Chen M, Au GG, Paradkar PN, Peiris M, Chung AW, Bloom JD, Lye D, Lok S, Wang LF
Sci Adv (2023) 9 pp. eade3470-eade3470 [ DOI: doi:10.1126/sciadv.ade3470 Pubmed: 37494438 ]
- Fab e7 heavy chain (23 kDa, Protein from Homo sapiens)
- Fab e7 light chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (132 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike glycoprotein trimer complexed with fab fragment of anti-rbd antibody e7 (Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Closed state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8d55
Q-score: 0.434
One RBD-up state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8d56
Q-score: 0.438