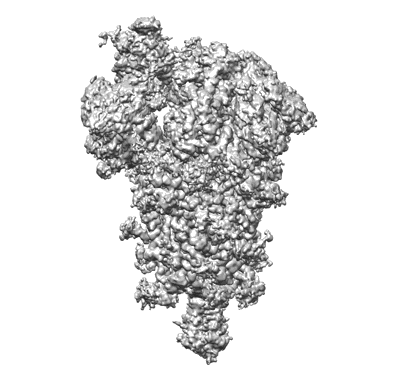
Closed state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8d55
Q-score: 0.434
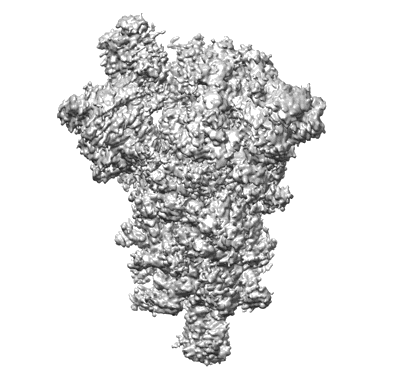
One RBD-up state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8d56
Q-score: 0.438
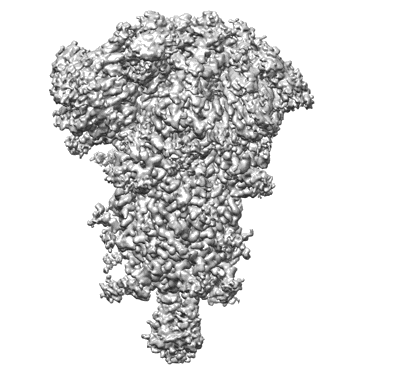
Middle state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8d5a
Q-score: 0.414
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Resolution: 2.9 Å
EM Method: Single-particle
Fitted PDBs: 8fdw
Q-score: 0.327
Shi W, Cai Y, Zhu H, Peng H, Voyer J, Rits-Volloch S, Cao H, Mayer ML, Song K, Xu C, Lu J, Zhang J, Chen B
Nature (2023) 619 pp. 403-409 [ Pubmed: 37285872 DOI: doi:10.1038/s41586-023-06273-4 ]
- Spike protein s2 (68 kDa, Protein from Severe acute respiratory syndrome coronavirus)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sras-cov-2 postfusion spike protein in nanodisc (210 kDa, Complex from Severe acute respiratory syndrome coronavirus 2)
- Alpha-d-mannopyranose (180 Da, Ligand)
cryo-EM structure of hemoglobin
Resolution: 3.5 Å
EM Method: Single-particle
Fitted PDBs: 7xgy
Q-score: 0.407
Zheng L, Liu N, Gao X, Zhu W, Liu K, Wu C, Yan R, Zhang J, Gao X, Yao Y, Deng B, Xu J, Lu Y, Liu Z, Li M, Wei X, Wang HW, Peng H
Nat Methods (2023) 20 pp. 123-130 [ DOI: doi:10.1038/s41592-022-01693-y Pubmed: 36522503 ]
- Protoporphyrin ix containing fe (616 Da, Ligand)
- Hemoglobin (Complex from Homo sapiens)
- Hemoglobin subunit beta (16 kDa, Protein from Homo sapiens)
- Hemoglobin subunit alpha (15 kDa, Protein from Homo sapiens)
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7tnw
Q-score: 0.44
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7to4
Q-score: 0.394
One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7sbl
Q-score: 0.394
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]
One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7sbt
Q-score: 0.269
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]
Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7sbk
Q-score: 0.434
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ DOI: doi:10.1126/science.abl9463 Pubmed: 34698504 ]