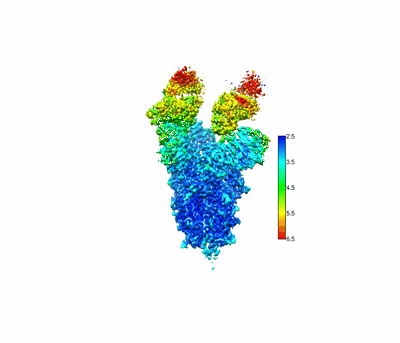
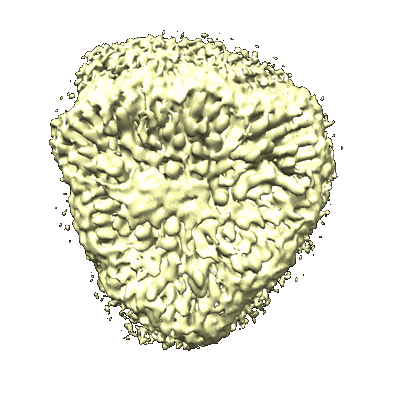
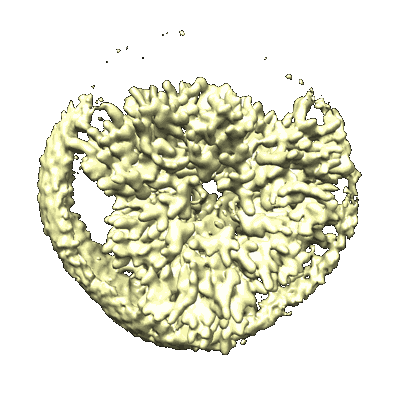
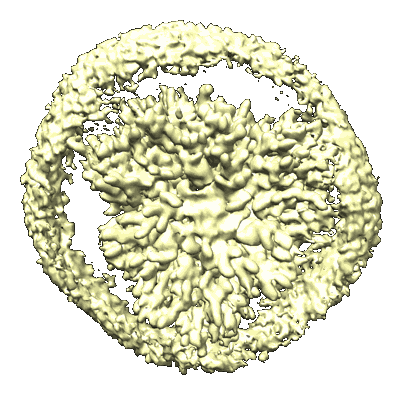
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8khc
Q-score: 0.351
Wang Y, Zhang Z, Yang M, Xiong X, Yan Q, Cao L, Wei P, Zhang Y, Zhang L, Lv K, Chen J, Liu X, Zhao X, Xiao J, Zhang S, Zhu A, Gan M, Zhang J, Cai R, Zhuo J, Zhang Y, Rao H, Qu B, Zhang Y, Chen L, Dai J, Cheng L, Hu Q, Chen Y, Lv H, So RTY, Peiris M, Zhao J, Liu X, Mok CKP, Wang X, Zhao J
Cell Rep (2024) 43 pp. 113653-113653 [ DOI: doi:10.1016/j.celrep.2023.113653 Pubmed: 38175758 ]
- Light chain of 5817 fab (11 kDa, Protein from Homo sapiens)
- Water (18 Da, Ligand)
- Spike glycoprotein (124 kDa, Protein from Severe acute respiratory syndrome coronavirus 2)
- Sars-cov-2 omicron spike in complex with 5817 fab (Complex from Severe acute respiratory syndrome coronavirus 2)
- Heavy chain of 5817 fab (13 kDa, Protein from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Resolution: 3.34 Å
EM Method: Single-particle
Fitted PDBs: 8iyx
Q-score: 0.448
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
- Cryo-em structure of the gpr34 receptor in complex with the antagonist yl-365 (Complex from Homo sapiens)
- 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid (583 Da, Ligand)
- Probable g-protein coupled receptor 34,probable g-protein coupled receptor 34,yl-365 (66 kDa, Protein from Homo sapiens)
Cryo-EM structure of GPR34-Gi complex
Resolution: 3.27 Å
EM Method: Single-particle
Fitted PDBs: 8sai
Q-score: 0.471
Xia A, Yong X, Zhang C, Lin G, Jia G, Zhao C, Wang X, Hao Y, Wang Y, Zhou P, Yang X, Deng Y, Wu C, Chen Y, Zhu J, Tang X, Liu J, Zhang S, Zhang J, Xu Z, Hu Q, Zhao J, Yue Y, Yan W, Su Z, Wei Y, Zhou R, Dong H, Shao Z, Yang S
PNAS (2023) 120 pp. e2308435120-e2308435120 [ Pubmed: 37733739 DOI: doi:10.1073/pnas.2308435120 ]
- Guanine nucleotide-binding protein g(i) subunit alpha-1 (40 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2 (7 kDa, Protein from Homo sapiens)
- Scfv16 (Complex from Mus musculus)
- Scfv16 (28 kDa, Protein from Mus musculus)
- Soluble cytochrome b562,probable g-protein coupled receptor 34, guanine nucleotide-binding protein g(i)/g(s)/g(o) subunit gamma-2, guanine nucleotide-binding protein g(i) subunit alpha-1, guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (Complex from Homo sapiens)
- Human gpr34 in complex with gi heterotrimer and scfv16 (Complex)
- Probable g-protein coupled receptor 34 (53 kDa, Protein from Homo sapiens)
- Guanine nucleotide-binding protein g(i)/g(s)/g(t) subunit beta-1 (39 kDa, Protein from Homo sapiens)
- O-[(s)-hydroxy{[(2s)-2-hydroxy-3-(octadec-9-enoyloxy)propyl]oxy}phosphoryl]-l-serine (523 Da, Ligand)
Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value
Resolution: 2.59 Å
EM Method: Single-particle
Fitted PDBs: 8iwx
Q-score: 0.466
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- Lhcii in detergent solution at high ph value (120 kDa, Complex from Spinacia oleracea)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Cryo-EM structure of protonated LHCII in detergent solution at low pH value
Resolution: 2.68 Å
EM Method: Single-particle
Fitted PDBs: 8iwy
Q-score: 0.572
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
- Chlorophyll b (907 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value
Resolution: 2.52 Å
EM Method: Single-particle
Fitted PDBs: 8iwz
Q-score: 0.563
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Lhcii in detergent solution at low ph value (120 kDa, Complex from Spinacia oleracea)
- Chlorophyll b (907 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Cryo-EM structure of unprotonated LHCII nanodisc at high pH value
Resolution: 2.64 Å
EM Method: Single-particle
Fitted PDBs: 8ix0
Q-score: 0.541
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Chlorophyll b (907 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- Lhcii nanodsic at high ph value (120 kDa, Complex from Spinacia oleracea)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Cryo-EM structure of protonated LHCII nanodisc at low pH value
Resolution: 2.63 Å
EM Method: Single-particle
Fitted PDBs: 8ix1
Q-score: 0.526
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
- Chlorophyll b (907 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Cryo-EM structure of unprotonated LHCII nanodisc at low pH value
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8ix2
Q-score: 0.531
Ruan M, Li H, Zhang Y, Zhao R, Zhang J, Wang Y, Gao J, Wang Z, Wang Y, Sun D, Ding W, Weng Y
Nat Plants (2023) 9 pp. 1547-1557 [ DOI: doi:10.1038/s41477-023-01500-2 Pubmed: 37653340 ]
- Chlorophyll a-b binding protein, chloroplastic (23 kDa, Protein from Spinacia oleracea)
- (3r,3'r,6s)-4,5-didehydro-5,6-dihydro-beta,beta-carotene-3,3'-diol (568 Da, Ligand)
- Lhcii nanodsic at low ph value (120 kDa, Complex from Spinacia)
- Chlorophyll b (907 Da, Ligand)
- (1r,3r)-6-{(3e,5e,7e,9e,11e,13e,15e,17e)-18-[(1s,4r,6r)-4-hydroxy-2,2,6-trimethyl-7-oxabicyclo[4.1.0]hept-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaenylidene}-1,5,5-trimethylcyclohexane-1,3-diol (600 Da, Ligand)
- (3s,5r,6s,3's,5'r,6's)-5,6,5',6'-diepoxy-5,6,5',6'- tetrahydro-beta,beta-carotene-3,3'-diol (600 Da, Ligand)
- 1,2-dipalmitoyl-phosphatidyl-glycerole (722 Da, Ligand)
- Chlorophyll a (893 Da, Ligand)
Resolution: 3.6 Å
EM Method: Single-particle
Fitted PDBs: 7yff
Q-score: 0.445
Zhang J, Zhang M, Wang Q, Wen H, Liu Z, Wang F, Wang Y, Yao F, Song N, Kou Z, Li Y, Guo F, Zhu S
Nat Struct Mol Biol (2023) 30 pp. 629-639 [ DOI: doi:10.1038/s41594-023-00959-z Pubmed: 36959261 ]
- (2r)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid (252 Da, Ligand)
- Glycine (75 Da, Ligand)
- Glutamate receptor ionotropic, nmda 1 (94 kDa, Protein from Homo sapiens)
- Glutamate receptor ionotropic, nmda 2d (97 kDa, Protein from Homo sapiens)
- Nmda receptor with nmda 1 incorperated with nmda 2d (384 MDa, Complex from Homo sapiens)
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)