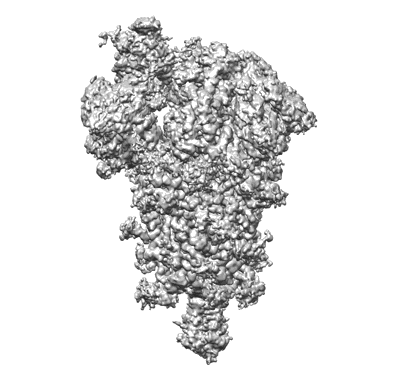
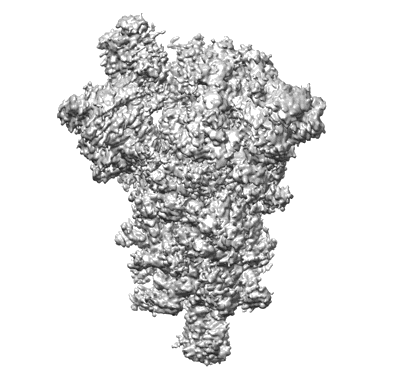
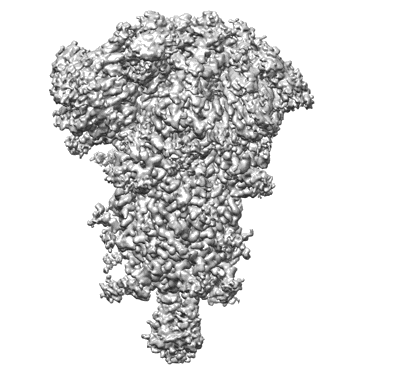
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Resolution: 3.12 Å
EM Method: Single-particle
Fitted PDBs: 7y71
Q-score: 0.415
Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Kwek MSS, Greaney AJ, Chen M, Au GG, Paradkar PN, Peiris M, Chung AW, Bloom JD, Lye D, Lok S, Wang LF
Sci Adv (2023) 9 pp. eade3470-eade3470 [ DOI: doi:10.1126/sciadv.ade3470 Pubmed: 37494438 ]
- 2-acetamido-2-deoxy-beta-d-glucopyranose (221 Da, Ligand)
- Sars-cov-2 spike glycoprotein trimer complexed with fab fragment of anti-rbd antibody e7 (Complex from Homo sapiens)
- Fab e7 heavy chain (23 kDa, Protein from Homo sapiens)
- Spike glycoprotein (132 kDa, Protein from Homo sapiens)
- Fab e7 light chain (23 kDa, Protein from Homo sapiens)
Resolution: 4.03 Å
EM Method: Single-particle
Fitted PDBs: 7y72
Q-score: 0.395
Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Kwek MSS, Greaney AJ, Chen M, Au GG, Paradkar PN, Peiris M, Chung AW, Bloom JD, Lye D, Lok S, Wang LF
Sci Adv (2023) 9 pp. eade3470-eade3470 [ DOI: doi:10.1126/sciadv.ade3470 Pubmed: 37494438 ]
- Fab e7 heavy chain (23 kDa, Protein from Homo sapiens)
- Sars-cov-2 spike glycoprotein trimer complexed with fab fragment of anti-rbd antibody e7 (Complex from Homo sapiens)
- Spike glycoprotein (132 kDa, Protein from Homo sapiens)
- Fab e7 light chain (23 kDa, Protein from Homo sapiens)
Closed state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 2.8 Å
EM Method: Single-particle
Fitted PDBs: 8d55
Q-score: 0.434
One RBD-up state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 3.0 Å
EM Method: Single-particle
Fitted PDBs: 8d56
Q-score: 0.438
Middle state of SARS-CoV-2 BA.2 variant spike protein
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 8d5a
Q-score: 0.414
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7tnw
Q-score: 0.44
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7to4
Q-score: 0.394
One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein
Resolution: 3.4 Å
EM Method: Single-particle
Fitted PDBs: 7sbl
Q-score: 0.394
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ Pubmed: 34698504 DOI: doi:10.1126/science.abl9463 ]
One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein
Resolution: 4.4 Å
EM Method: Single-particle
Fitted PDBs: 7sbt
Q-score: 0.269
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ Pubmed: 34698504 DOI: doi:10.1126/science.abl9463 ]
Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein
Resolution: 3.1 Å
EM Method: Single-particle
Fitted PDBs: 7sbk
Q-score: 0.434
Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, Tong P, Gautam A, Mayer ML, Walsh Jr RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu J, Chen B
Science (2021) 374 pp. 1353-1360 [ Pubmed: 34698504 DOI: doi:10.1126/science.abl9463 ]