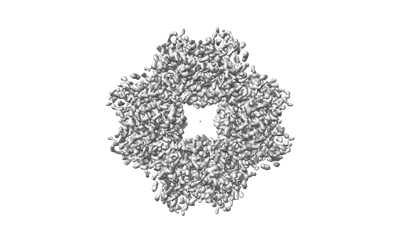
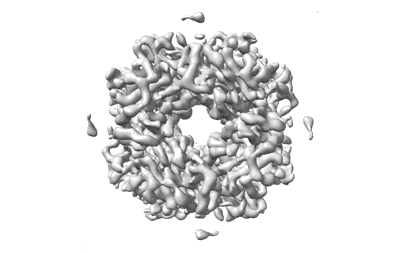
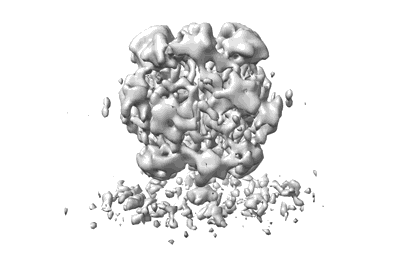
Cryo-ET structure of RuBisCO from 3.9 angstroms Synechococcus elongatus PCC 7942
Resolution: 3.9 Å
EM Method: Subtomogram averaging
Fitted PDBs: 8wpz
Q-score: 0.444
Kong WW, Zhu Y, Zhao HR, Du K, Zhou RQ, Li B, Yang F, Hou P, Huang XH, Chen Y, Wang YC, Sun F, Jiang YL, Zhou CZ
Structure (2024) 32 pp. 1110-1120.e4 [ DOI: doi:10.1016/j.str.2024.05.007 Pubmed: 38823379 ]
- Ribulose bisphosphate carboxylase small subunit (13 kDa, Protein from Synechococcus elongatus PCC 7942 = FACHB-805)
- Ribulose bisphosphate carboxylase large chain (52 kDa, Protein from Synechococcus elongatus PCC 7942 = FACHB-805)
- Rubisco (Complex from Synechococcus elongatus (strain ATCC 33912 / PCC 7942 / FACHB-805))
Cryo-ET map of RuBisCO at 4.4 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome
Cryo-ET map of RuBisCO-SSUL at 5.9 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome
Resolution: 3.66 Å
EM Method: Single-particle
Fitted PDBs: 8hdr
Q-score: 0.445
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ Pubmed: 36656854 DOI: doi:10.1073/pnas.2213727120 ]
- Pam3 sheath protein (41 kDa, Protein from uncultured cyanophage)
- Pam3 connector protein (12 kDa, Protein from uncultured cyanophage)
- Pam3 tube protein (15 kDa, Protein from uncultured cyanophage)
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 terminator protein (17 kDa, Protein from uncultured cyanophage)
- Pam3 adaptor protein (13 kDa, Protein from uncultured cyanophage)
Resolution: 3.96 Å
EM Method: Single-particle
Fitted PDBs: 7yfw
Q-score: 0.413
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ Pubmed: 36656854 DOI: doi:10.1073/pnas.2213727120 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 fiber proreins (27 kDa, Protein from uncultured cyanophage)
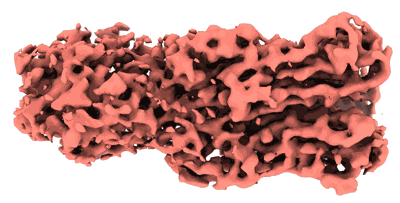
Cyanophage Pam3 baseplate proteins
Resolution: 3.19 Å
EM Method: Single-particle
Fitted PDBs: 7yfz
Q-score: 0.528
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ Pubmed: 36656854 DOI: doi:10.1073/pnas.2213727120 ]
- Pam3 plug gp18 (11 kDa, Protein from uncultured cyanophage)
- Pam3 baseplate wedge gp22 (32 kDa, Protein from uncultured cyanophage)
- Pam3 hub gp19 (25 kDa, Protein from uncultured cyanophage)
- Pam3 spike gp20 (23 kDa, Protein from uncultured cyanophage)
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 baseplate wedge gp23 (26 kDa, Protein from uncultured cyanophage)
- Pam3 sheath initiator gp21 (13 kDa, Protein from uncultured cyanophage)
- Pam3 tube initiator gp17 (19 kDa, Protein from uncultured cyanophage)
Cyanophage Pam3 portal-adaptor
Resolution: 3.57 Å
EM Method: Single-particle
Fitted PDBs: 8hds
Q-score: 0.47
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ Pubmed: 36656854 DOI: doi:10.1073/pnas.2213727120 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Pam3 portal protein (69 kDa, Protein from uncultured cyanophage)
- Pam3 adaptor protein (13 kDa, Protein from uncultured cyanophage)
Cyanophage Pam3 capsid asymmetric unit
Resolution: 3.17 Å
EM Method: Single-particle
Fitted PDBs: 8hdt
Q-score: 0.537
Yang F, Jiang YL, Zhang JT, Zhu J, Du K, Yu RC, Wei ZL, Kong WW, Cui N, Li WF, Chen Y, Li Q, Zhou CZ
PNAS (2023) 120 pp. e2213727120-e2213727120 [ Pubmed: 36656854 DOI: doi:10.1073/pnas.2213727120 ]
- Uncultured cyanophage (Virus from uncultured cyanophage)
- Major capsid (34 kDa, Protein from uncultured cyanophage)
- Cement (17 kDa, Protein from uncultured cyanophage)